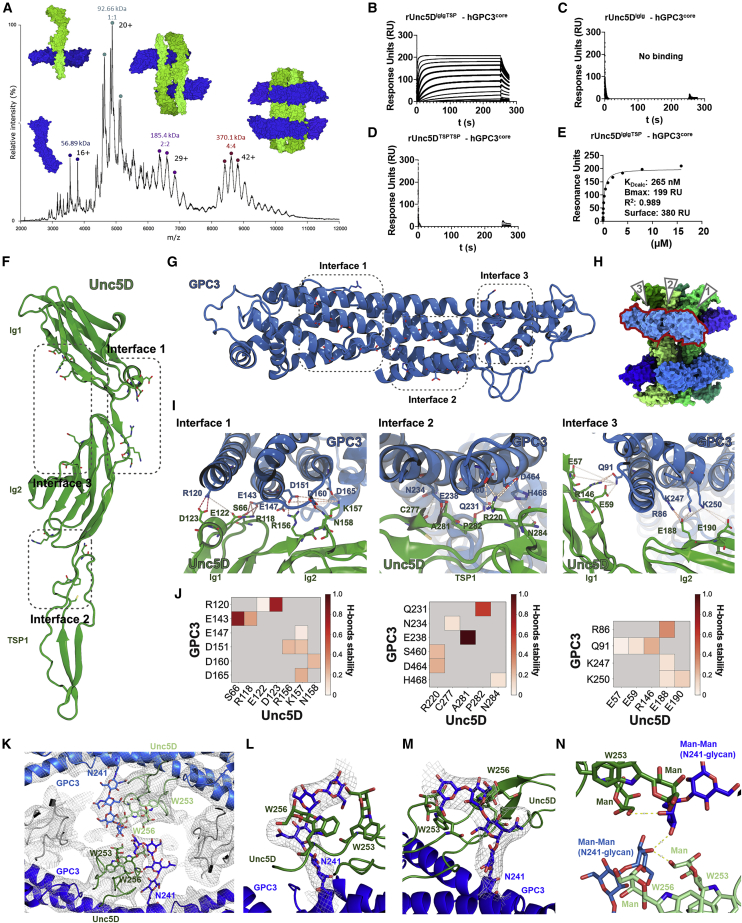
Figure 2.
Characterization of the hGPC3-rUnc5 complex
(A) Native MS spectrum of rUnc5IgIgTSP and hGPC3core (R355A/R358A). Charge state series (labeled with colored dots) are assigned to the complexes shown. Individual peaks were isolated for MS/MS analysis to identify subcomplexes (Figures S1F–S1H).
(B–E) SPR data shows binding that rUnc5IgIgTSP, but not the shorter constructs rUnc5DIgIg and rUnc5DTSPTSP, binds hGPC3core with nanomolar affinity. The apparent KD (KDcalc) was calculated using a 1:1 binding model and is indicative only.
(F) Binding interfaces 1–3 on rUnc5DIgIgTSP.
(G) Binding interfaces 1–3 on hGPC3core.
(H) Binding interfaces 1–3 indicated on the octameric complex. The glypican molecule for which these are shown is outlined in red.
(I) Zoomed views of interacting residues in interfaces 1–3 (hGPC3core-rUnc5DIgIgTSP complex). Hydrogen bonds are shown as dotted yellow lines.
(J) Summaries of the hydrogen bond analyses during restrained molecular dynamics (MD) simulation. Atoms that contribute to stable hydrogen bonds between the two proteins are shown, and colored blocks indicate the stability of the bond during simulation (averages for the four copies of the complex). Non-averaged results are shown in Figures S2A–S2C.
(K) View of the glycan emanating from two copies of hGPC3 N241 toward the center of the complex. C-mannosylated tryptophans of nearby rUnc5D TSP1 domains are indicated (W253, W256). The calculated 2FoFc map of the hGPC3-rUnc5D complex data is shown as a gray mesh (sigma = 1).
(L and M) As (K), but showing zoomed views of the N241-glycan for one of the hGPC3 copies within the complex. The map is carved around the N-linked glycan.
(N) Distances below 3.5 Å between atoms within glycans from different chains are indicated as yellow dotted lines.
See also Figure S2.