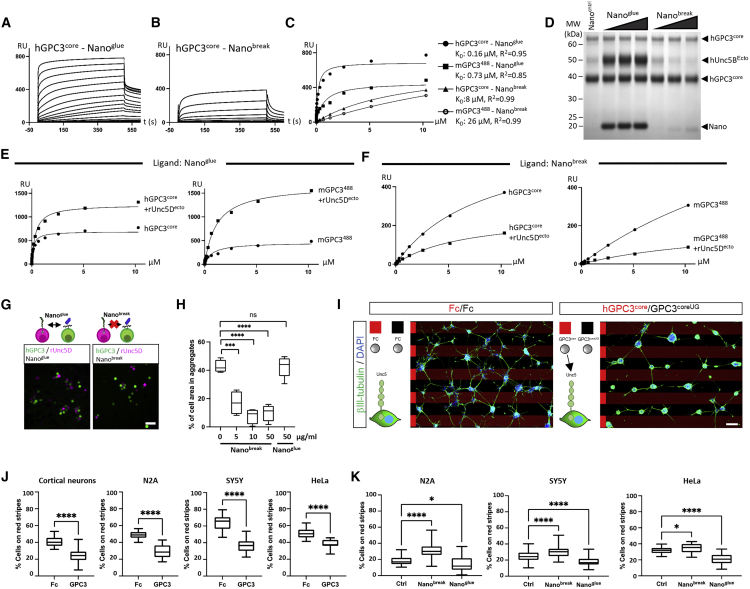
Figure 4.
Nanobodies enhance or disrupt GPC3-Unc5 interaction
(A) SPR binding data using hGPC3core and Nanoglue.
(B) As in (A), with Nanobreak.
(C) The equilibrium values from experiments shown in panels A and B, and equivalent values from using mGPC3488 (Figures S4E and S4F). KD values were calculated assuming 1:1 binding.
(D) Unc5B pull downs with immobilized hGPC3core using Nanoglue and Nanobreak.
(E and F) Equilibrium SPR data (raw data in Figures S4E and S4F) confirms that Nanoglue enhances, and Nanobreak weakens, GPC3-Unc5D binding.
(G) Cell-cell aggregation assay using soluble Nanobreak and Nanoglue. Representative images.
(H) Quantification of cell-cell aggregation experiments.
(I) E15.5 dissociated cortical neurons were grown on alternate stripes (red and black) containing Fc, GPC3core, or GPC3coreUG.
(J) Quantification beta-III-tubulin+ (green) pixels on red stripes: Fc (=Fc/Fc control), GPC3 (=hGPC3core/hGPC3coreUG). We performed equivalent stripe assays also for HeLa, N2A and SY5Y cells, using DAPI for quantification. ∗∗∗∗p < 0.0001, Student T-tests.
(K) Results from stripe assays in the presence of streptavidin (Ctrl) or streptavidin-nanobody complexes (Nanoglue or Nanobreak).
We performed one-way ANOVA with Tukey’s post hoc tests (H) and (K). NS, not significant; ∗p < 0.05; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. Scale bars represent 100 μm (G) and 90μm (I).
See also Figure S4.