Figure S1.
mGPC3core structure and GPC3-Unc5D complex data, related to Figure 1
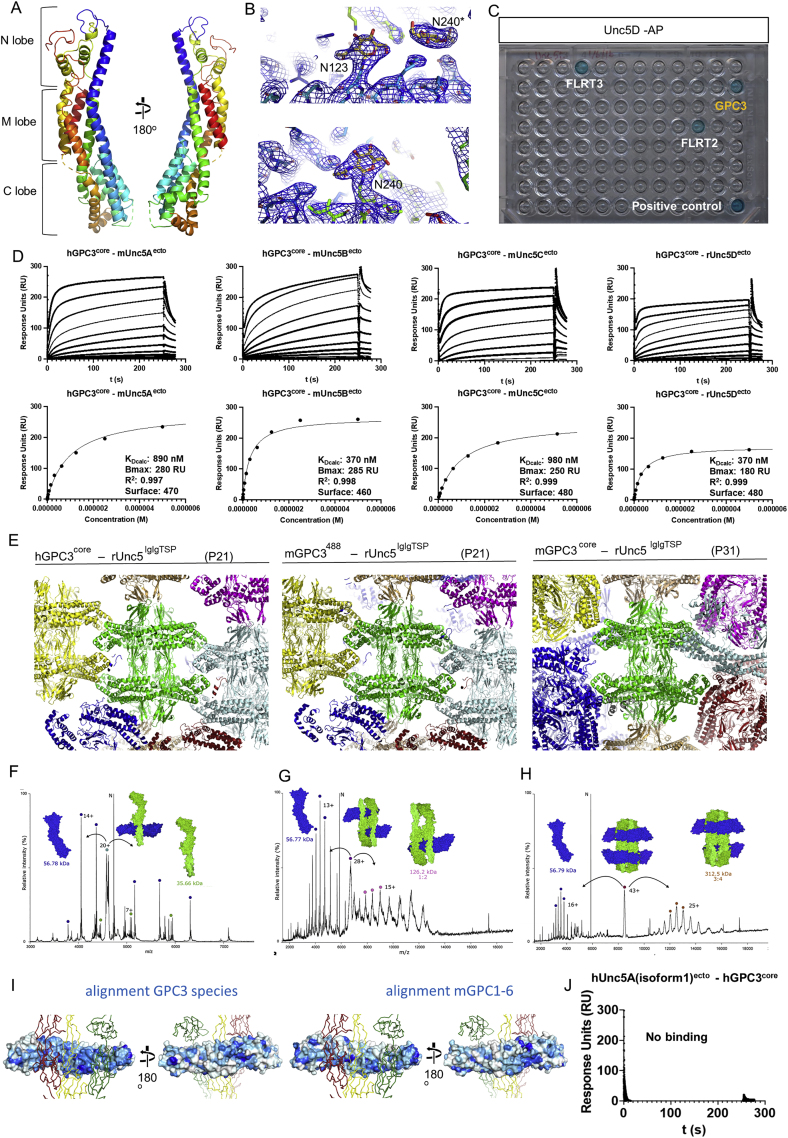
(A) Mouse GPC3core structure colored according to the rainbow (blue: N-terminus, red: C-terminus).
(B) Electron density map calculated from murine GPC3core crystals is shown in blue, centered on N123 and N240 of a symmetry-related molecule∗ (left) and N240 (right).
(C) ELISA plate contained Unc5D-AP (human, residues 33–379) immobilized in each well as bait and 95 other Fc-tagged proteins were added as prey, as described in (Ozgul et al., 2019). As expected, FLRT2/3 bind to Unc5D. GPC3 (human, residues 25–563) was a new positive interactor. Positive control was NRXN1-Fc/NLGN1-AP as described in (Ozgul et al., 2019).
(D) SPR experiments show binding of Unc5 extracellular domains to hGPC3core. The apparent KDs (KDcalc) were calculated using a 1:1 binding model and are indicative only. Bmax, R2 and amount of ligand immobilised on the flowcell surface are indicated.
(E) Crystal packing environment for the three complex structures. Each octameric unit is shown in a different color, with a central unit in green.
(F–H) Tandem MS (MS/MS) analysis of peaks presented in Figure 2A. Peaks reveal rUnc5IgIgTSP and hGPC3core(R355A/R358A) subcomplexes. The 93 kDa peak dissociated into masses corresponding to GPC3 (56.78 kDa excluding glycans) and Unc5D (35.66 kDa excluding glycans), the peaks corresponding to 185 and 370 kDa dissociated into GPC3 (56.78 kDa) and a mass of 126 kDa (consistent with a 2:1 Unc5D:GPC3 complex). In the 370 kDa peak we additionally detected a 312 kDa species (consistent with a 4:3 Unc5D:GPC3 complex). Charge state series (labeled with colored dots) are assigned to the complexes shown.
(I) rUnc5DIgIgTSP is shown in red, yellow and green ribbons, as found in the complex with mGPC3core. The surface of mGPC3core is colored in shades of blue according to sequence conservation (blue = conserved, while = not conserved). Surface conservation was calculated using aligned sequences from human, mouse, opossum, chicken, frog, and fish GPC3 (top) or mouse GPC1-6 (bottom). Note that the Unc5-binding site is less conserved amongst mouse GPC1-6 sequences, compared to different GPC3 sequences.
(J) SPR results show that hUnc5A isoform A, which lacks a TSP1 domain, is unable to bind hGPC3core.