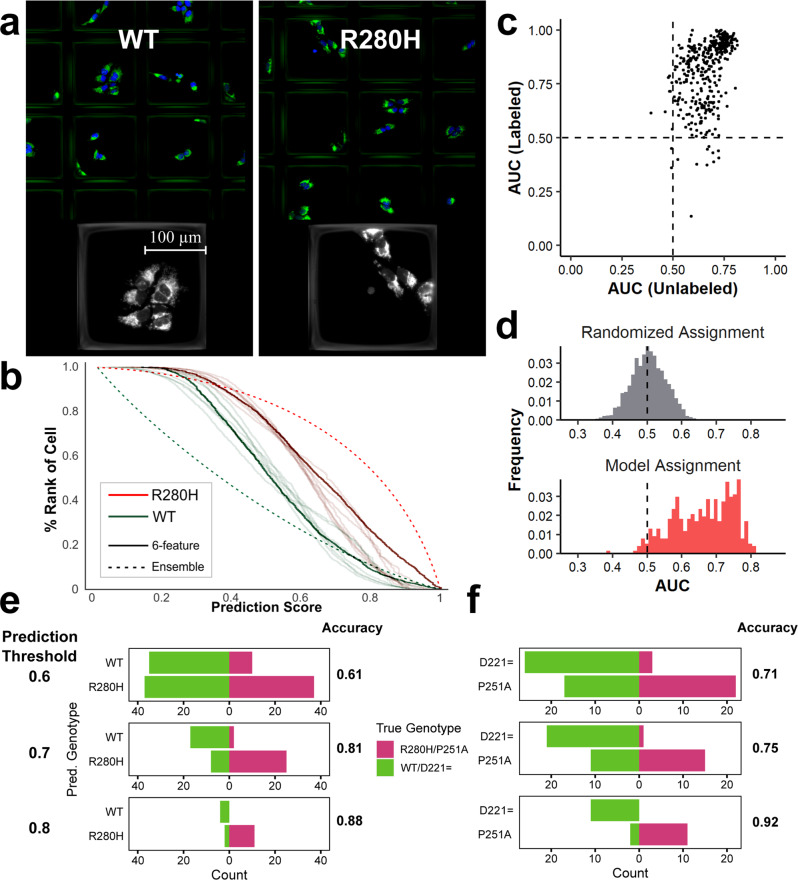
Fig. 4. Raft-seq can predict MFN2 WT and MFN2 R280H mutant cells which have nearly undetectable phenotypic differences.
a Images of the R280H mutant and MFN2 WT cell line in the microraft plate. b Ranked histogram of the prediction scores for several models predicting the genotype from the R280H experiment. Each curve shows the distribution of prediction scores for a particular labeled genotype (WT or R280H) where a score of 0 means the model is confident that a particular cell is WT and a score of 1 means the model has a strong prediction of R280H. 8 different 6-feature models are shown. The dashed lines indicate an ensemble model. c Scatterplot showing the performance of all 433 models in detecting mutants in a mixture of wild-type cells and the R280H mutant. d A histogram of AUCs for models detecting mutants in a mixture of wild-type cells and the R280H mutant. A histogram of AUCs generated from randomly assigned models for comparison. e, f Accuracies and confusion bar charts of the predictions from the unlabeled wells, when only picking cells with prediction scores ≥0.9 (≤0.1), ≥0.8 (≤0.2), and ≥0.7 (≤0.3). e uses the model to predict R280H against WT MFN2, while f uses the model to predict P251A against D221=.