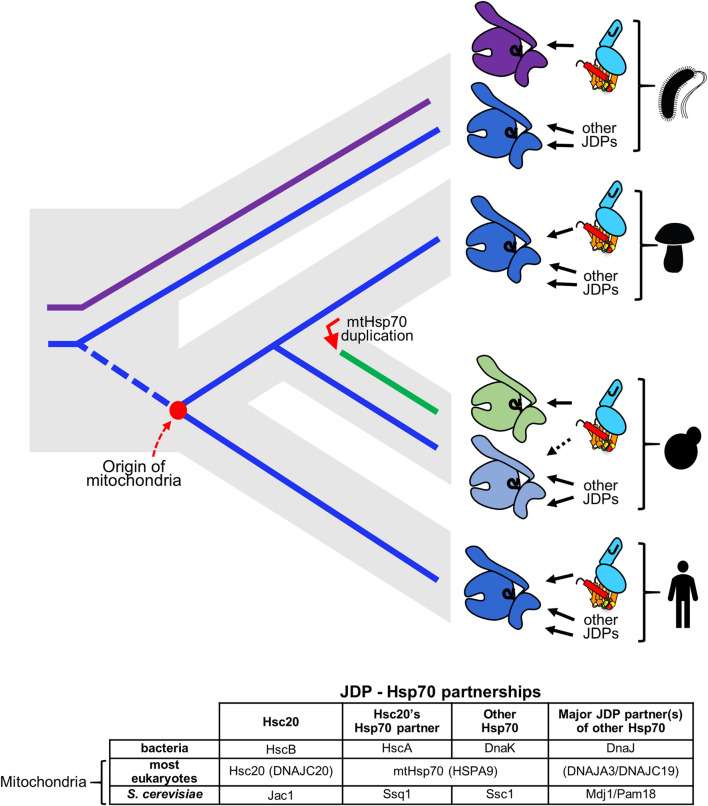
FIGURE 4.
Hsc20 has changed Hsp70 partners twice across the tree of life. (top) Gray contour indicates phylogenetic relationships among evolving lineages: bacteria, fungi, metazoa. Solid lines indicate evolution of Hsp70s HscA (magenta) and DnaK (blue); broken blue line indicates the eukaryotic lineage prior to the origin of mitochondria (red dot). JDP Hsc20 is found in bacteria and mitochondria of eukaryotes (cyan, shown bound to client Isu/IscU in brown with FeS cluster). Solid arrows indicate its Hsp70 partner. In bacteria Hsc20 (HscB) partners with FeS cluster specialized Hsp70 HscA (magenta). This system coexists with multifunctional Hsp70 DnaK (dark blue). In mitochondria of eukaryotic cells Hsc20 partners with mtHsp70 e.g., most fungi and metazoans (indicated by mushroom and human pictograms) or with FeS cluster specialized Hsp70 Ssq1 (green), which evolved after an Hsp70 gene duplication (red arrow) in a progenitor of the S. cerevisiae lineage (budding yeast pictogram). The other duplicate, Ssc1 (light blue) remained multifunctional partnering with multiple JDPs, but only minimally with Hsc20, as indicated by dotted line. Following the origin of mitochondria Hsc20 was maintained, but HscA was not. Its function in FeS cluster biogenesis was substituted by mtHsp70, a descendant of DnaK (dark blue). (bottom) Names of JDPs and Hsp70s functioning in FeS cluster biogenesis. Those in parentheses are the name of human proteins.