FIGURE 5.
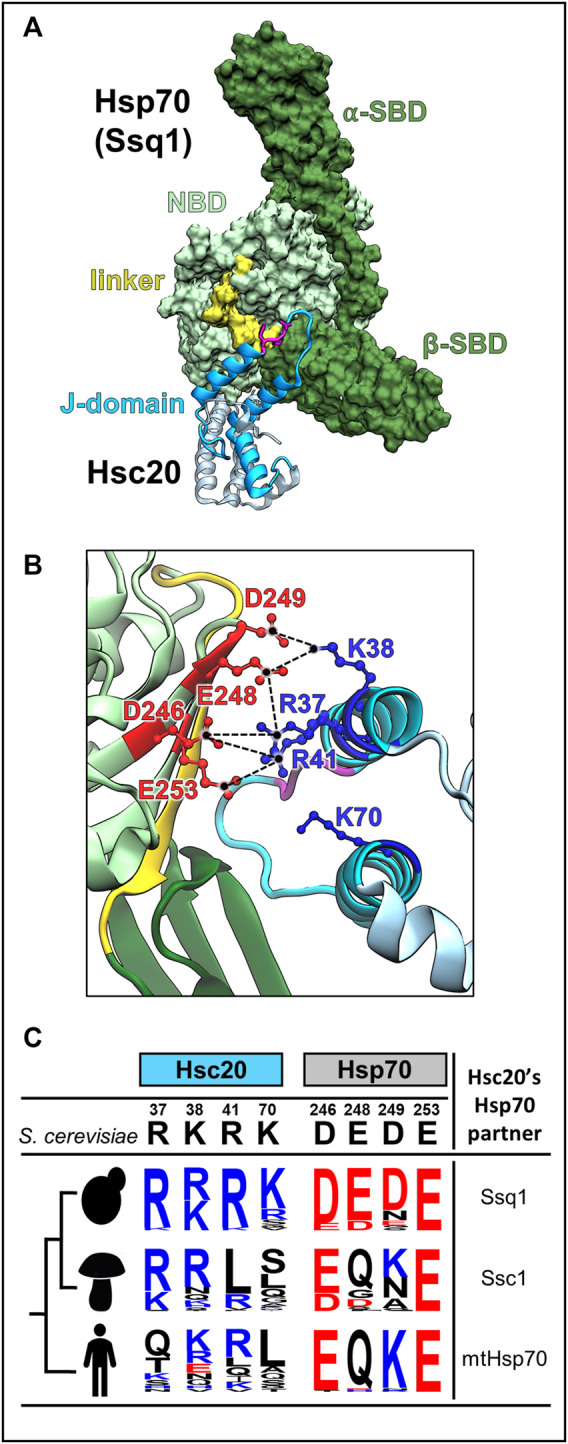
J-domain-Hsp70 interaction involves evolutionary variable interface. (A) Structural model of the S. cerevisiae Hsc20-Ssq1 (ATP) complex (Tomiczek et al., 2020). As in all JDP-Hsp70 interactions, the J-domain binds at the interface of NBD, β-SBD and interdomain linker formed when Ssq1 is in the ATP-bound conformation. Hsc20 in cyan with J-domain dark, Isu binding domain, light; HPD motif (magenta). Ssq1: NBD (light green), α- and β-SBDs (dark green); interdomain linker (yellow) is bound to the NBD. (B) The interconnected network of contacts between positively charged residues of helix 2 of the J-domain (blue) and negatively charged residues of Ssq1 (red) is critical for J-domain interaction with Ssq1; electrostatic interactions are depicted as broken lines (black). Note that one residue on the J-domain side interacts with more than one residue on the Hsp70 side e.g., J-domain’s R41 contacts D246 and E253 of Ssq1. K70 of helix 3 is also critical for Hsc20-Ssq1 complex formation, though its binding partner(s) was not identified. (C) The key residues involved in the J-domain-Ssq1 interaction are evolutionary variable. Sequence logos represent the amino acid frequency at positions of Hsc20 and Hsp70, which are homologous to the hot spot positions of the S. cerevisiae Hsc20 J-domain-Ssq1 interface; positively charged (blue), negatively charged (red) and uncharged (black). The analysis was based on Hsc20 and Hsp70 sequences from 23 yeast species, which diverged after Hsp70 gene duplication leading to Ssq1, 47 pre-duplication fungi species (harboring only mtHsp70) and 20 metazoan species (harboring mtHsp70), designated by human figure. Note: in contrast to the invariant HPD motif hot spot residues involved in J-domain/Ssq1 interaction in S. cerevisiae are evolutionary variable, particularly outside the post-duplication species.
