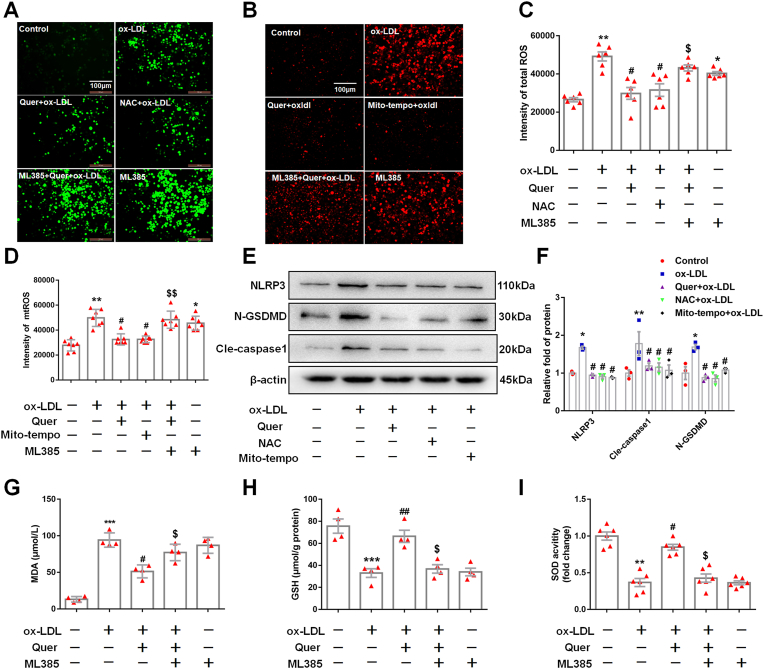
Fig. 5.
ML385 abolishes the anti-ROS effect of quercetin.
5 μM ML385 was pretreated in 1 h, then 50 μM quercetin is added into the medium, after 1 h, 100 μg/ml ox-LDL is also added into the medium for 24 h. Immunofluorescence is used to observed the ROS staining, microplate reader is used to evaluated the intensity of ROS. The macrophages are divided into six groups, control groups, ox-LDL groups (ox-LDL, 100 μg/ml), Quer group (pretreated with 50 μM quercetin for 1 h, then combined with 100 μg/ml ox-LDL for 24 h), NAC group (pretreated with 10 μM NAC for 1 h, then combined with 100 μg/ml ox-LDL for 24 h), Mito-tempo group (pretreated with 10 μM mito-tempo for 1 h, then combined with 100 μg/ml ox-LDL for 24 h), ML385 group (5 μM ML385). Western blot is used to analyze the proteins of NLRP3, N-GSDMD and cle-caspase1. (A). Representative images for cellular ROS. (B). Representative images for mtROS. (C). Quantized data for cellular ROS (n = 6). (D). Quantized data for mitochondrial ROS (n = 7). (E, F). The expression of NLRP3, N-GSDMD, and cle-caspase1 (n = 3). (G). MDA level in control, ox-LDL, Quer + ox-LDL, ML385 (5 μM) + Quer + ox-LDL and ML385 (5 μM) group (n = 4). (H). GSH level in control, ox-LDL, Quer + ox-LDL, ML385 (5 μM) + Quer + ox-LDL and ML3853 (5 μM) group (n = 4). (I). SOD activity in control, ox-LDL, Quer + ox-LDL, ML3853 (5 μM) + Quer + ox-LDL and ML3853 (5 μM) group (n = 6). *p < 0.05, **p < 0.01, ***p < 0.01 vs control group. #p < 0.05, vs ox-LDL group. $p < 0.05, $$p < 0.01 vs Quer + ox-LDL group.