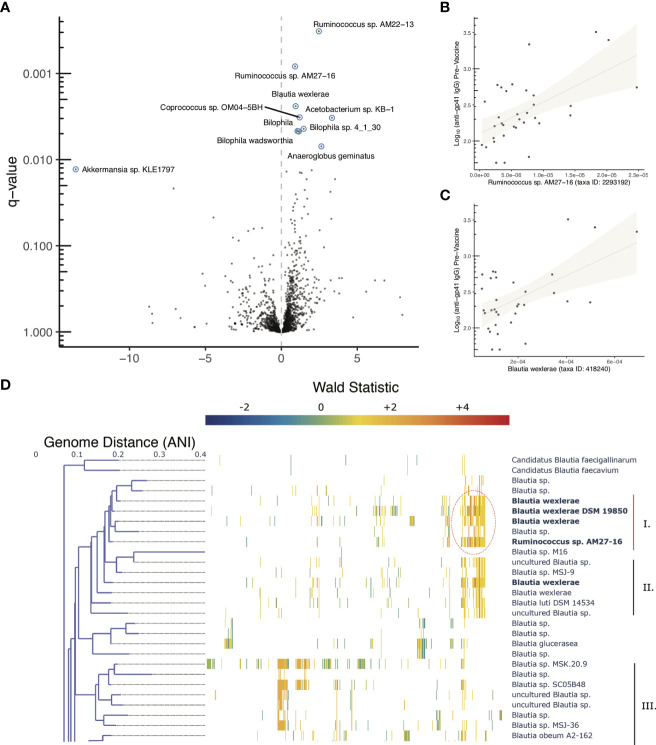
Figure 5.
Microbiome associations with pre-vaccine gp41 IgG and Clustered co-abundant genes (CAGs) from fecal samples collected in HVTN 106. (A) Volcano plot of taxonomic features (the sum of all genes identified as part of any Clustered co-abundant genes (CAGs) assigned to a taxa, (B, C) Relative abundance of two of the most statistically significant taxa (x-axis) compared with pre-vaccine anti-gp41IgG (y-axis). (D) Fine scale mapping of genes identified in CAGs to genomes in the Blautia genus. Gene’s are mapped to genomes if they share 90% alignment identity at the amino acid level. The genes belonging to CAGs most consistently associated with anti-gp41 IgG pre vaccine have higher positive Wald statistics. Figure S3 shows the full tree, containing members of the genus lacking genes associated with anti-gp41 IgG.