Figure 1.
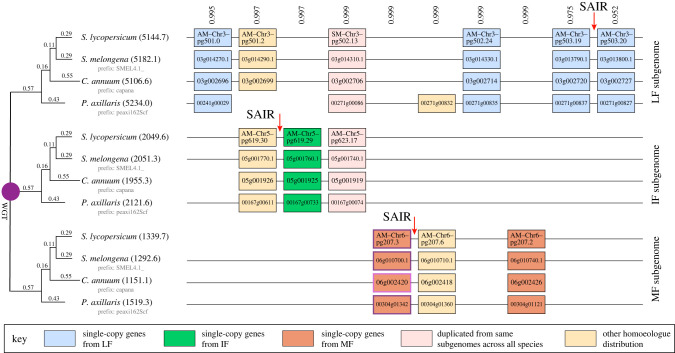
Subgenome assignment and inference of gene loss after the shared hexaploid in four Solanaceae species, showing our inferences of an LF subgenome and two more fractionated ones (IF and MF). The tree at the left illustrates POInT's estimates of the homoeologue loss rates at each stage of the evolution of these four hexaploid genomes. POInT's estimated branch length (αt) for each branch is shown above it. This length is used by the model to compute the probability of a gene that is triplicated at the start of the branch experiencing one or more gene losses along that branch. In parentheses, after each taxa are the extant number of genes that POInT estimates survive from that subgenome in that particular modern genome. Shown at the right is an example window of 16 loci produced by whole genome triplication (WGT). Each column (or pillar) corresponds to an ancestral locus triplicated through the hexaploidy, with the boxes representing extant genes. Pairs of genes are connected by lines if they are genomic neighbours in the modern genome. The numbers above each pillar are the posterior probabilities assigned to these combinations of orthology relationships related to the other 64-1 possible orthology states. Three subgenome-assigned intergenic regions (SAIRs) are illustrated (see Methods). (Online version in colour.)