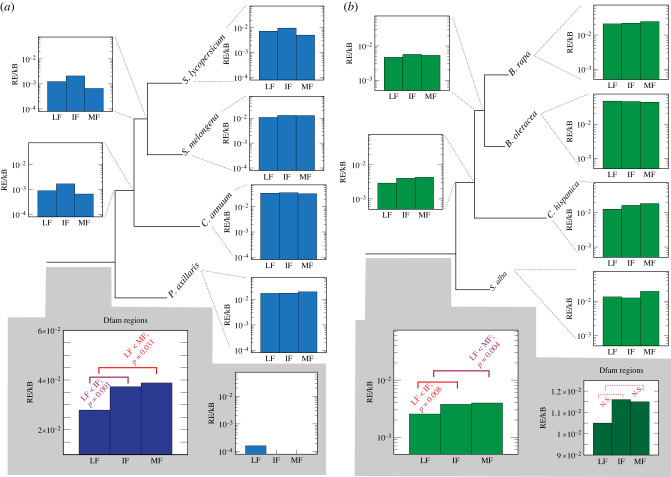
Figure 3.
Independent and shared RE density in SAIRs. For each extant genome at the phylogeny tips (right of the trees), we show the repeat density (RE/kB) for each subgenome, inferred with BLASTN and RepBase (Methods). At the phylogeny roots (grey shaded regions) and internal branches (left of the tree), we show the density of repeats that are found in orthologous SAIRs for the species in that clade (e.g. for the root branch, all four species in the tree). Note the log-scale on the y-axis of all diagrams except that for Dfam, reporting RE hits per kilobase (kB) of SAIR sequence. For the Brassiceae, there are significantly fewer such insertions shared between all species in the LF genome (see Methods), but this difference is not seen for the Solanaceae genomes. For the Solanaceae, LF is significantly deficient in shared REs when the larger Dfam set of elements is used as a query, but the differences, while in the same direction for the Brassiceae, are non-significant (Methods). (Online version in colour.)