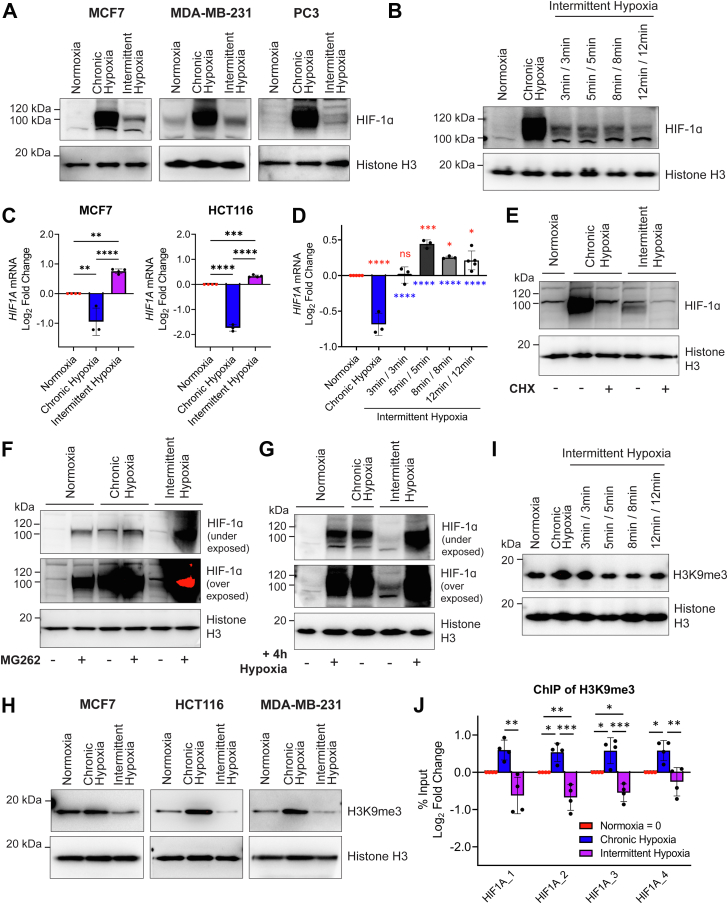
Figure 1.
Intermittent hypoxia increases HIF1A expression, decreases global H3K9me3, and decreases H3K9me3 at the HIF1A gene locus.A, nuclear HIF-1α levels in MCF7, MDA-MB-231, and PC3 cells exposed to normoxia, chronic hypoxia, and intermittent hypoxia (5 min/5 min). B, nuclear HIF-1α levels in MCF7 cells exposed to normoxia, chronic hypoxia, and varying periods of intermittent hypoxia. C, HIF1A mRNA levels in MCF7 and HCT116 cells exposed to normoxia, chronic hypoxia, and intermittent hypoxia (5 min/5 min). D, HIF1A mRNA levels in MCF7 cells exposed to normoxia, chronic hypoxia, and varying periods of intermittent hypoxia. Mean ± S.D. of n ≥ 3 experiments. Values are normalized to normoxia (Log2 scale). E, nuclear HIF-1α in HCT116 cells exposed to normoxia, chronic hypoxia, and intermittent hypoxia (5 min/5 min) treated with DMSO or cycloheximide (CHX). F, nuclear HIF-1α in HCT116 cells exposed to normoxia, chronic hypoxia, and intermittent hypoxia (5 min/5 min) with DMSO or MG262. G, nuclear HIF-1α in MCF7 cells exposed to 18 h normoxia, chronic hypoxia, and intermittent hypoxia (5 min/5 min) with or without an additional 4 h of chronic hypoxia. H, H3K9me3 levels in HCT116, MCF7, and MDA-MB-231 cells exposed to normoxia, chronic hypoxia, and intermittent hypoxia (5 min/5 min). I, H3K9me3 levels in MCF7 cells exposed to normoxia, chronic hypoxia, and varying periods of intermittent hypoxia. Histone H3 is used as a loading control for (A, B and E). J, chromatin immunoprecipitation was performed in MCF7 cells for H3K9me3 and enrichment on four different sites along the HIF1A gene relative to the total amount of input chromatin was quantified by qPCR. Values are normalized to normoxia (Log2 scale). Mean ± SD of n = 4. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Asterisks above a line compare data points connected by the line. Red asterisks above the data compare that condition to normoxia. Blue asterisks below the data compare that condition to chronic hypoxia. DMSO, dimethyl sulfoxide; qPCR, quantitative PCR.