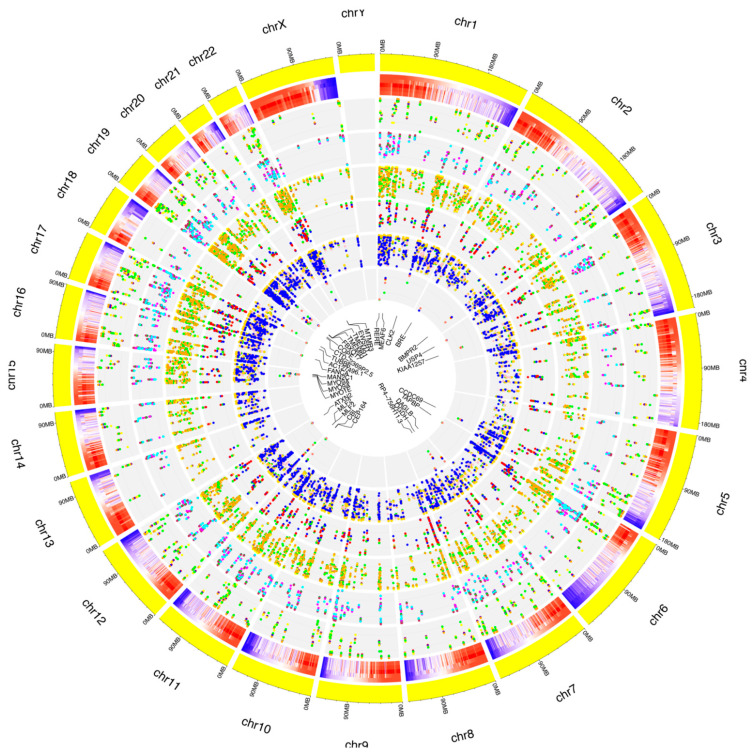
Figure 5.
Distribution of known RNA editing sites from RPE cells across human chromosomes. The human genome is shown as a circle. For each chromosome, the position of the editing sites along with their average editing ratio (heatmap) is shown for different time points. From outside to inside: (1) N° of A-to-I only editing sites common to all considered time points and controls. The red, blue, orange, yellow and green dots indicate CTRL, 1 h, 2 h, 3 h and 6 h time points, respectively. (2) N° of C-to-U only editing sites common to all considered time points and controls. The purple, green, maroon, magenta and cyan dots indicate CTRL, 1 h, 2 h, 3 h and 6 h time points, respectively. (3) N° of editing sites within other-than-CDS and UTRs common to all considered time points and controls. The coral, navy, yellow, green and orange dots indicate CTRL, 1 h, 2 h, 3 h and 6 h time points, respectively. (4) N° of editing sites within CDS common to all considered time points and controls. The yellow, salmon, green, blue and red dots indicate CTRL, 1 h, 2 h, 3 h and 6 h time points, respectively. (5) N° of editing sites within UTRs common to all considered time points and controls. The green, orange, gold, pink and blue dots indicate CTRL, 1 h, 2 h, 3 h and 6 h time points, respectively. (6) N° of editing sites within repetitive elements common to all considered time points and controls. The yellow, red, blue, green and salmon dots indicate CTRL, 1 h, 2 h, 3 h and 6 h time points, respectively. For these editing sites, the related gene names are indicated.