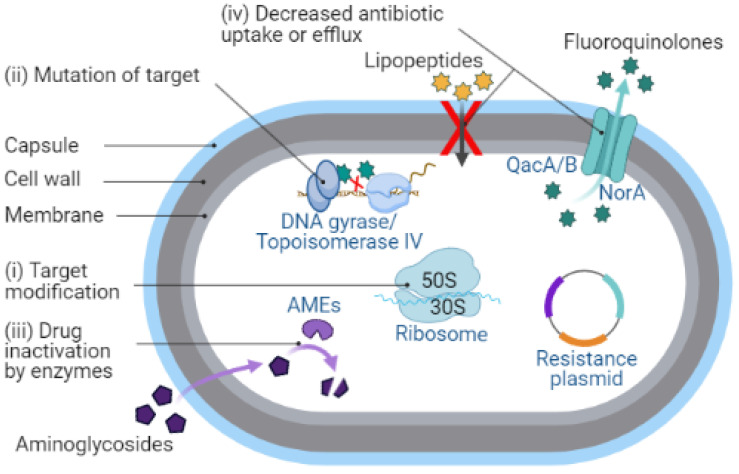
Figure 2.
Schematic representation of the mechanisms of antibiotic resistance in S. aureus. (i) Target modification: change in the structure or composition of the target site in a bacterial cell can stop the antibiotic to bind, thus shielding it from the antibiotic. Modification of the bacterial ribosome by 23S ribosomal RNA (rRNA) methyltransferase (encoded by erm genes) leads to a conformational change in the ribosome target [31,32], thereby preventing the binding of erythromycin to the ribosomal target. (ii) Mutation of target: mutations in the target can prevent the antibiotic from binding, or if it binds, preventing it from inhibiting the target. Mutation in the DNA topoisomerase IV subunit A (encoded by grlA gene) and an essential enzyme DNA gyrase subunit A (encoded by gyrA gene) is the main contributor to fluoroquinolone resistance in S. aureus [33,34,35]. (iii) Drug inactivation by enzymes: several S. aureus enzymes modify the structure of antibiotics or break them down to make them inactive. The bifunctional aminoglycoside-modifying enzyme (AME) AAC(6′)/APH(2″) (encoded by aac(6′)/aph(2″) genes) confers resistance to aminoglycosides via acetyltransferase and phosphotransferase activities [36,37]. (iv) Decreased antibiotic uptake or efflux: decrease in the permeability of cell membrane to drugs makes it more difficult to pass through or activation of an efflux pump that removes antibiotics from the bacterial cell. The norA, qacA/B, and smr (qacC/D) genes encoding multidrug efflux pump proteins are found mainly in S. aureus clinical isolates and mediate resistance to fluoroquinolones, tetracyclines, and reduced susceptibility to certain antiseptics [38,39].