Figure 1.
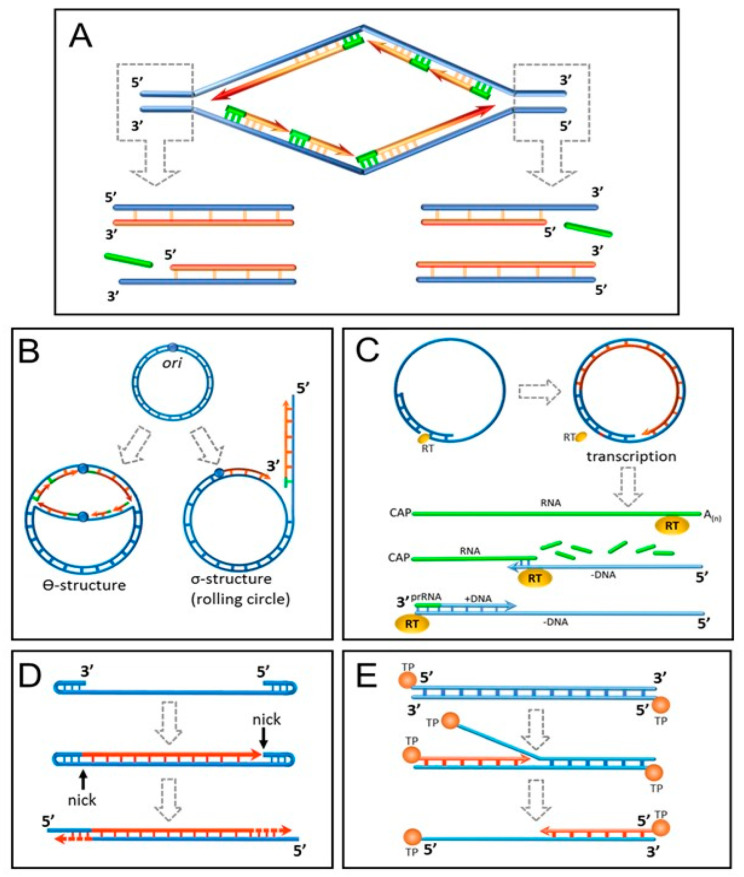
The problem of incomplete DNA replication and its solution by “primitive” organisms. (A) Incomplete replication of the 3′ end of DNA in eukaryotic cells. Replicons located in the telomere region cannot displace the last RNA primer of the lagging strand. (B) A covalently closed circular genome replication. In theta replication (prokaryotes, viruses), the last RNA primer of the lagging strand is replaced by the leading strand. A rolling cycle mechanism (viruses, plasmids) uses a single-stranded nick as a primer for DNA synthesis. As a result, a concatemer containing several complete genome copies is formed. (C) Hepadnaviruses do not have a covalently closed circular genome, but it can be easily restored by reparative DNA synthesis after cell infection. Despite this, the virus uses reverse transcription as a strategy for its genome replication. Whole genome RNA, after its translation, is used as a template for minus DNA strand synthesis. Reverse transcriptase (RT) includes a domain that can prime DNA synthesis from a tyrosine residue. The incomplete plus DNA strand is synthesized by DNA polymerase primed with the rest of the degraded whole genome RNA. (D) Parvoviruses use hairpins at the ends of their DNA (rabbit ears) as primers for DNA polymerase. (E) Adenovirus DNA polymerase forms a complex with a terminal protein (TP) that can act as a primer for unidirectional DNA replication.