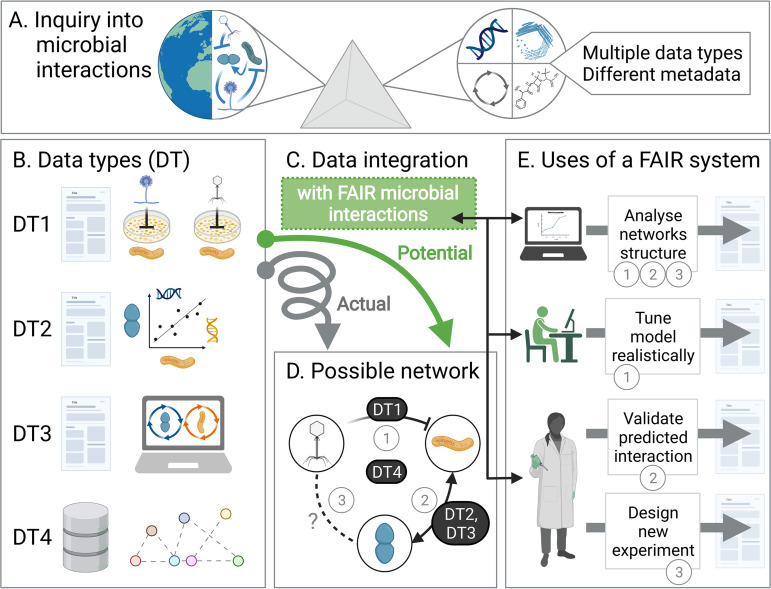
FIG 1.
Applying a FAIR system to the study of microbial interactions and correlations. (A–B) Studies that investigate microbial interactions gain relevant insights through the generation of multiple types of data and metadata (DTs). As representative examples of microbe–microbe interactions, we may consider fungi–bacteria or phage–bacteria cultivation experiments (DT1), correlations based on amplicon sequence or operational taxonomic unit counts (DT2), and flux balance models of genome-scale metabolic networks of two or more species (DT3). While specific databases (DT4 and Table 1) that compile these data sources exist, they lack a common reporting standard, which hinders downstream application and integration. (C) We envision a systematic approach for reporting microbial interactions following the principles of Findability, Accessibility, Interoperability, and Reusability (FAIR). (D) A FAIR representation of microbial interaction data, based on common identifiers for microorganisms and specific encodings of interactions and uncertainty, can enable new insights that bridge subdisciplines and generate predictions of new interaction networks. (E) Scientists spanning diverse areas of microbiome science can benefit from FAIR reporting of interactions. For example, a network scientist could identify common structures relying on a broader corpus of interactions, a modeler could more easily identify specific interactions to realistically simulate ecological dynamics, and an experimentalist could assess whether a novel interaction has been reported in other hosts or contexts. All these uses of the framework would lead not only to new scientific insights and more streamlined contribution to data collections, but also to growing interconnectedness within the diverse field of microbiome science.