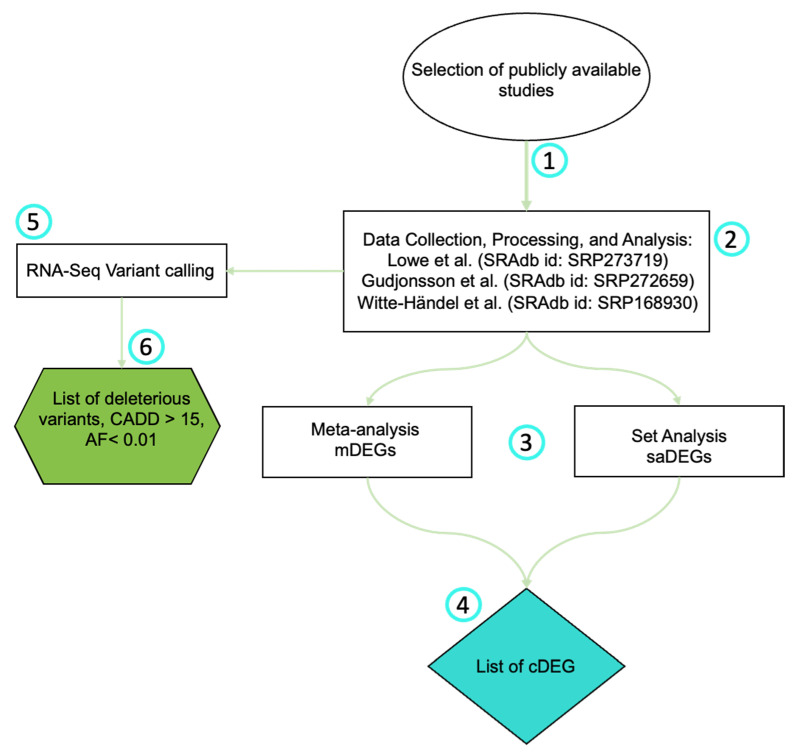
Figure 1.
Analysis flowchart: 1: Publicly available studies were selected from the Sequence Read Archive (SRA) and Gene Expression Omnibus (GEO); 2: Three studies passed our selection criteria and were downloaded. SRAdb package for R software version 4.1.0 was used to download all raw. fastq files. Trimmomatic v0.39 was used to trim Illumina adapters and to exclude reads. Resulting gene counts tables were converted into DESeq2 package objects [37,38,39]; 3: Lists of DEGs found through the meta-analysis (mDEGs) or the set analysis (saDEGs) (See supplementary data) were created; 4: List of common DEGs (cDEGs) found in both analyses was made (See supplementary data); 5: RNA-Seq variant calling analysis was performed by Strelka v2.9.10; 6: A list of potential HS-related variants originated based on deleterious capacity, allele frequency, and significant genotypic variation based on Wilcoxon Test (p value < 0.5).