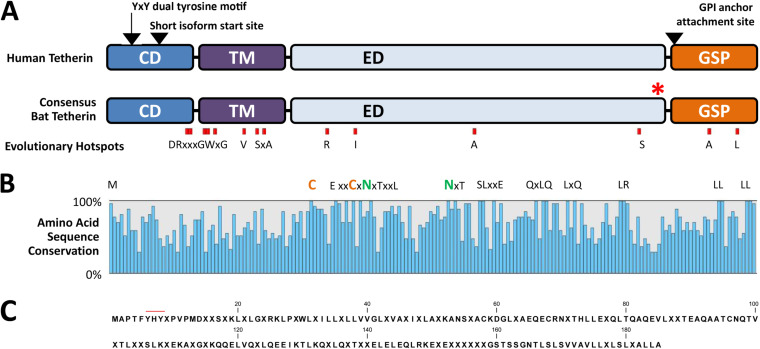
FIG 1.
Bat tetherin structure and sequence diversity. (A) Consensus bat tetherin amino acid sequence structures and motifs generated through the multiple sequence alignment of tetherin from 27 bat species (Data Set S1) and compared to human tetherin. Amino acid residues under strong positive selection are denoted by red boxes. The Y·X·Y dual-tyrosine motif, the alternative start site for the short isoform of tetherin, and the GPI-anchor attachment site positions are indicated by arrows. Red asterisk (*) indicates location of the inserted molecular tags in the Pteropus alecto and Myotis macropus tetherin expression constructs. CD, cytoplasmic domain; TM, transmembrane domain; ED, extracellular domain; GPI, glycophosphatidylinositol; GSP, GPI signal peptide. (B) Sequence diversity among bat tetherin proteins is indicated by the percentage of amino acid sequence conservation at each site of the consensus bat tetherin. Amino acids conserved in all 27 sequences are represented by their letters. Amino acids represented by X are variable and are included to indicate the sequence distances between closely positioned conserved residues. (C) Consensus bat tetherin sequence. Amino acid residues represented in >50% of bat tetherin sequences are indicated with their letter. Positions at which no amino acid residue is represented in >50% of bat tetherin sequences are indicated with ‘X’. The red line indicates the position of the dual-tyrosine motif.