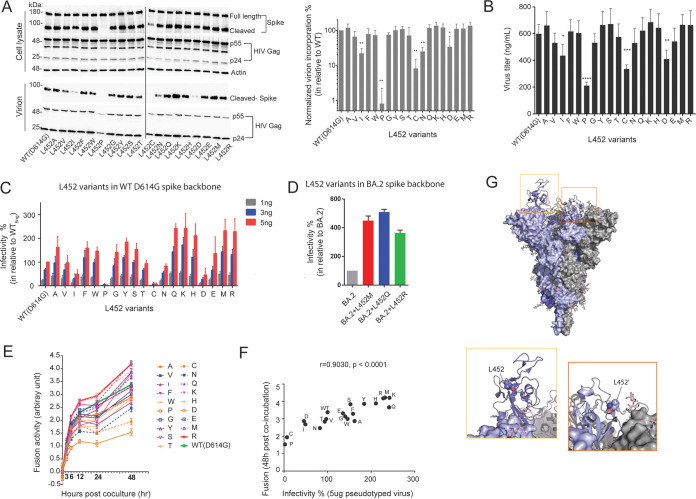
FIG 3.
Biochemical and virological characterization of spike L452 variants. (A) Immunoblots showing total cellular expression and virion incorporation of spike L452 mutants in producer cell and virion (left panel). The spike incorporation level was quantified and normalized to p24 Gag levels and is expressed relative to the WT in three independent replicates (right panel). Statistical significance was determined by one-way ANOVA with multiple-comparison tests (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (B) Titers (ng/mL) of pseudoviruses collected from the culture supernatant at 48 h posttransfection. (C) Infectivity of reporter lentiviruses pseudotyped by SARS-CoV-2 spike L452-mutants and WT D614G. The indicated titers of pseudoviruses (1, 3, and 5 ng) were exposed to 293T cells expressing ACE2 and TMPRSS2. The amount of pseudoviruses successfully infected into target cells was determined from the luciferase activity, and the relative infectivity is expressed as the percentage normalized to the WT (infection at 5 ng). (D) Infectivity of reporter lentiviruses pseudotyped with SARS-CoV-2 BA.2 spike and its L452 mutants (infection at 5 ng). (E) Fusion formation between the spike-expressing cells and ACE2-TMPRSS2-expressing cells was continuously monitored at intervals of 3, 6, 12, 24, and 48 h. The fusion activity was expressed relative to WT at 3 h after coincubation. (F) Correlation between infectivity of pseudoviruses (infection at 5 ng) and cell-to-cell fusion activity (at 48 h). The statistical significance was determined by using the Pearson’s correlation coefficient test. (G) Structure of SARS-CoV-2 spike trimer with single RBD up-conformation (PDB 7KRR). Each protomer is differently colored. The L452 residues are shown as spheres, while glycans are indicated as sticks. In the RBD with the up-conformation, the L452 residue is exposed (yellow box). In contrast, in the RBD with the down-conformation, the L452 residue is buried at the interface with the NTD of another protomer (orange box). The data shown are means ± the SD of triplicate determinations.