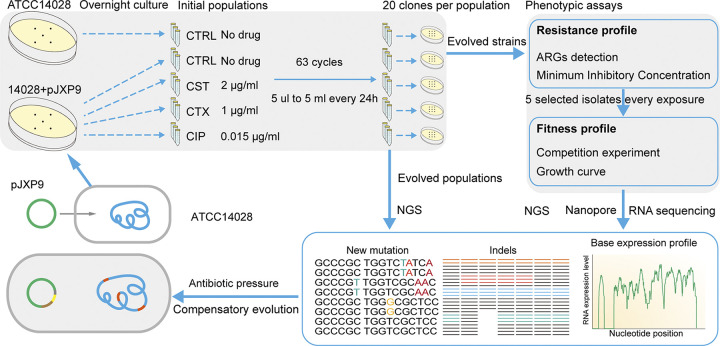
FIG 7.
Schematic overview for adaptive evolution of plasmid-bacterial host under subinhibitory or nondrug exposure. Colonies taken from LB agar plates of ancestral clones ATCC 14028+pJXP9 and ATCC 14028 were picked and inoculated into fresh LB broth as initial cultures and serially passaged in triplicate for 63 days. Twenty clones from each evolved population were selected for MICs and ARG detection, and representative clones (n = 5) were selected for estimating plasmid fitness effects by competition assays and growth curves. Evolved populations and evolved clones were further submitted to WGS by Illumina. Nanopore sequencing was also performed for representative clones harboring evolved IncHI2 plasmids of different sizes. Furthermore, evolved clones carrying plasmids of different sizes were subjected to whole-genome RNA sequencing. Parallel mutations for evolved populations and clones and mRNA abundance alterations were analyzed.