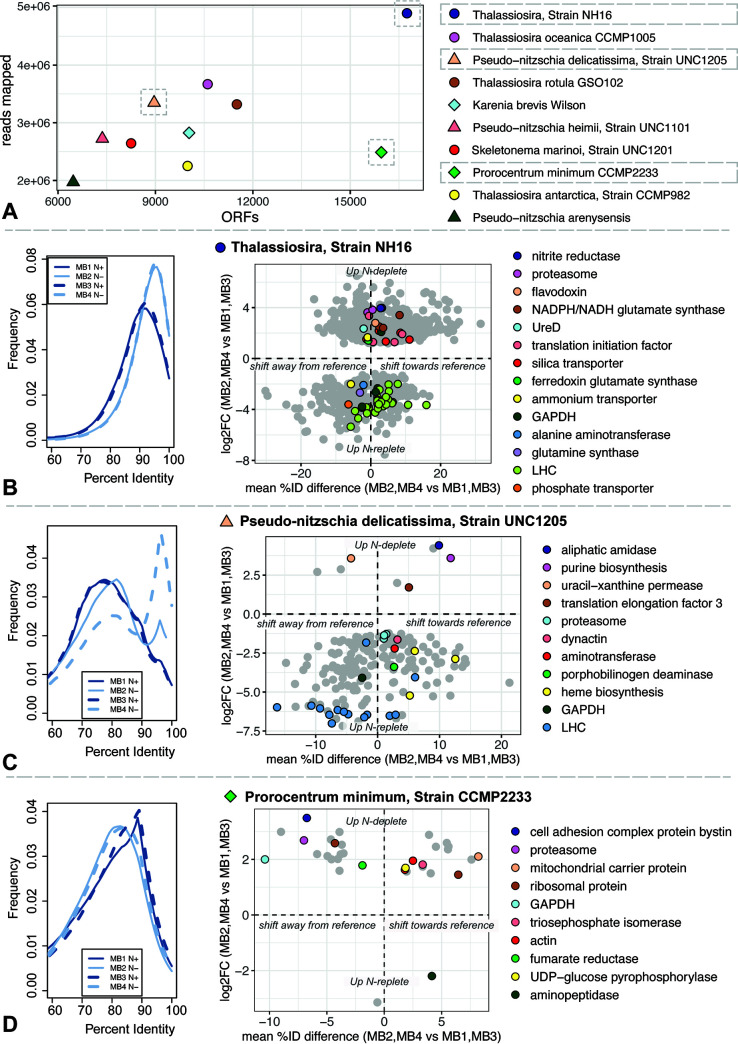
FIG 3.
Fine-scale taxonomic shifts across the nitrogen depletion experiment. (A) Reads (y axis) mapping to and number of ORFs (x axis) mapped to top 10 most abundantly hit Marine Microbial Eukaryotic Transcriptome Sequencing Project (MMETSP) reference transcriptomes. Circles, triangles, and diamonds represent centric diatom, pennate diatom, and dinoflagellate references, respectively. (B to D) References are indicated with dashed gray lines. References are the centric diatom Thalassiosira strain NH16 (B), the pennate diatom Pseudo-nitzschia delicatissima strain UNC1205 (C), and the dinoflagellate Prorocentrum minimum strain CCMP2233 (D). Left, percent identity histograms of reads mapped to MMETSP reference contigs for all nitrogen conditions. Right, log fold changes (y axis) versus mean percent identity differences (x axis) for reads mapping to reference ORFs across nitrogen conditions. Only ORFs that are significantly differentially expressed, at least 60% identical to the reference, and shift in the same direction relative to the reference across replicates are shown. ORFs of interest are colored by function.