FIG 9.
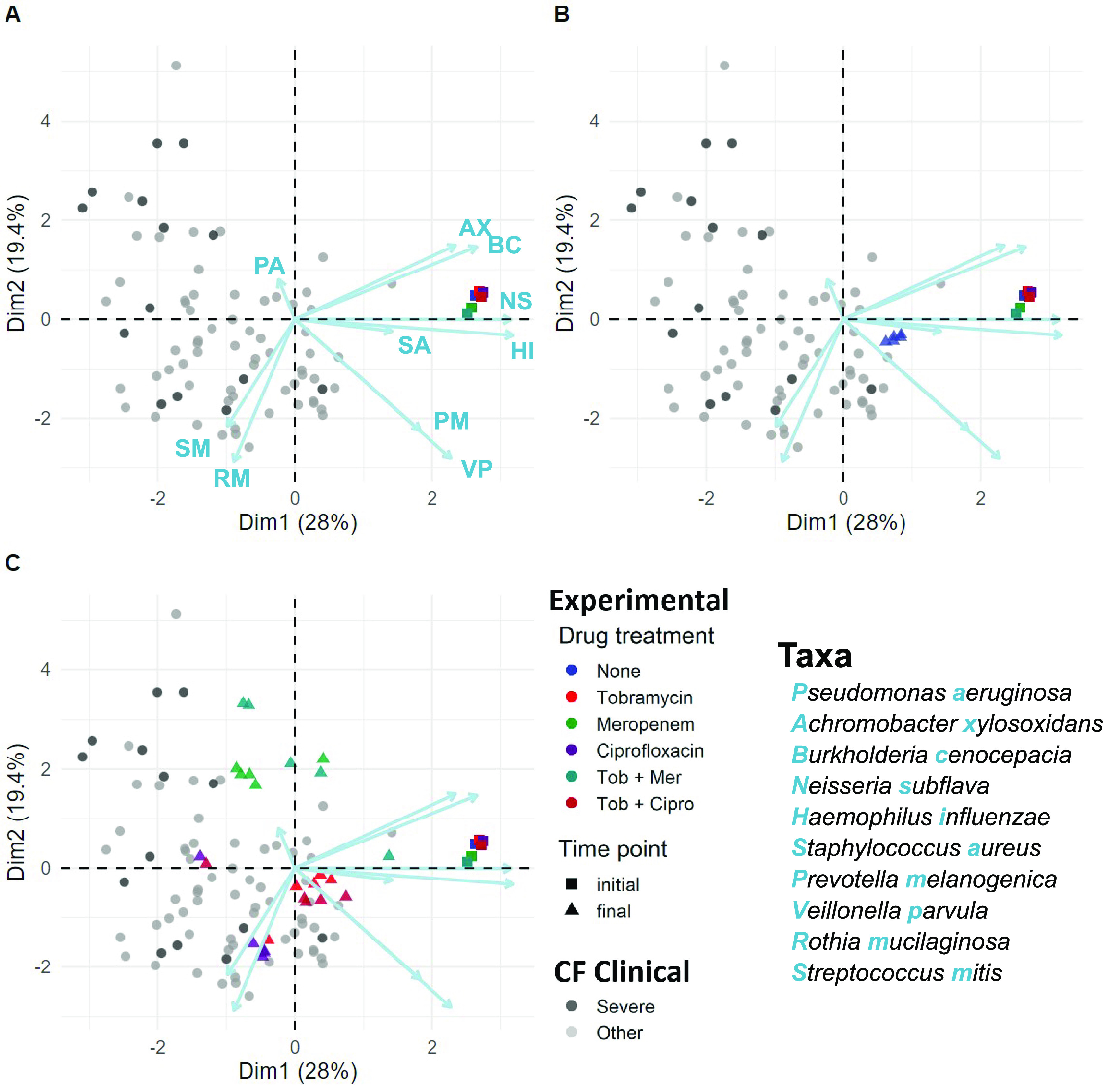
Antibiotics drive pathogen enrichment in experimental microbiomes, producing community structures that overlap clinical sputum communities. PCA visualization of experimental microbiome data (colored triangles and squares, summarizing data in Fig. 4) plus clinical microbiome data across a cohort of 77 people with CF (gray/black circles, black/severe signifies low lung function [57]). (A) Squares illustrate experimental initial conditions. (B and C) Triangles are final compositions after 5 serial passages (10 days), in the absence (B) or presence (C) of antibiotics. Colors denote experimental condition (see key). Each experimental treatment is replicated 5-fold, producing highly repeatable dynamics in the absence of antibiotics (blue triangles, B) and variable pathogen enriched outcomes following antibiotic treatment (C). Antibiotics were supplemented at each passage at clinically relevant concentrations (meropenem, 15 μg/mL; tobramycin, 5 μg/mL; ciprofloxacin, 2.5 μg/mL). Each point is a single microbiome sample (species resolution for clinical samples via the DADA2 plugin in QIIME2 [57, 119]). Ordination is the PCA of centered log-ratio transformed relative abundances.
