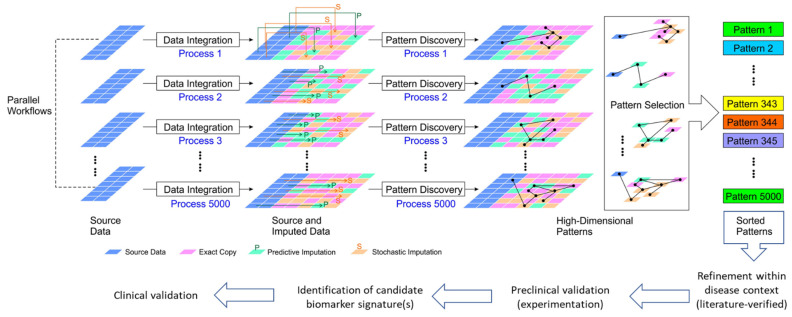
Figure 1.
Schematic illustration of our proposed very-large-scale predictive data integration and imputation process for high-dimensional pattern discovery as candidate biomarker signatures. The source parallel multi-omics data are amplified in multiple data imputation processes (see Methods). Predictive imputation uses the dependencies between data dimensions to fill in missing data and replace data with uncertainty in different configurations. Stochastic imputation further increases the variety of generated data by helping existing data to form more variety of patterns. Using this framework, we can expose more data patterns to the accessible operational range of existing pattern recognition algorithms. Data imputation tools provide more frequent and more diversified opportunities for pattern discovery [19]. Thus, the hidden patterns are placed at shallower, easier to discover data entry locations for many imputation instances. This very-large-scale data amplification and integration process allows us to boost existing pattern discovery tools to solve more challenging information dependencies as candidate integrated biomarker signatures for further validation.