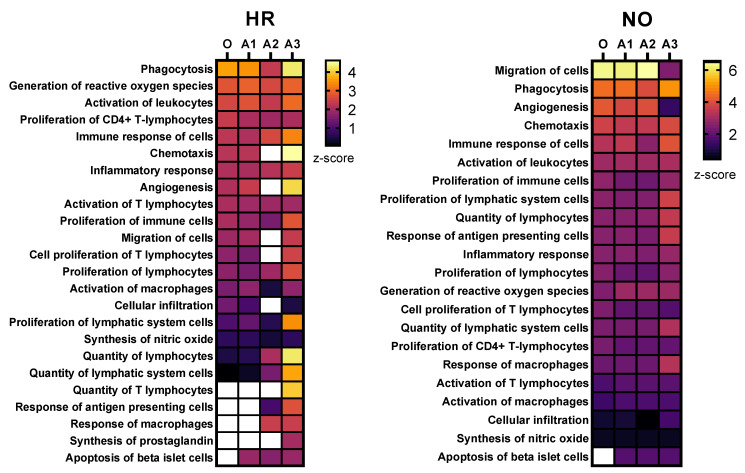
Figure 4.
Heat maps showing selected immune functions identified in the integrated amplified proteomics and metabolomics datasets (A1, A2, and A3) in comparison to the original data (O) for both the HR and NO subject groups. Heat maps were generated based on the average z-score assigned by IPA to each predicted function in the metabolomics and proteomics datasets for both amplified and original datasets. Functions were sorted by the z-score for the original dataset. The z-score is a statistical measure that accounts for the directional effect of change and the magnitude of its impact on the affected disease/function (https://go.qiagen.com/IPA-transcriptomics-whitepaper, accessed on 27 August 2022). Functions were selected based on their direct involvement in inflammatory and immune responses and, hence, as relevant to T1D (see Supplementary Tables S3 and S4 for complete lists of identified functions).