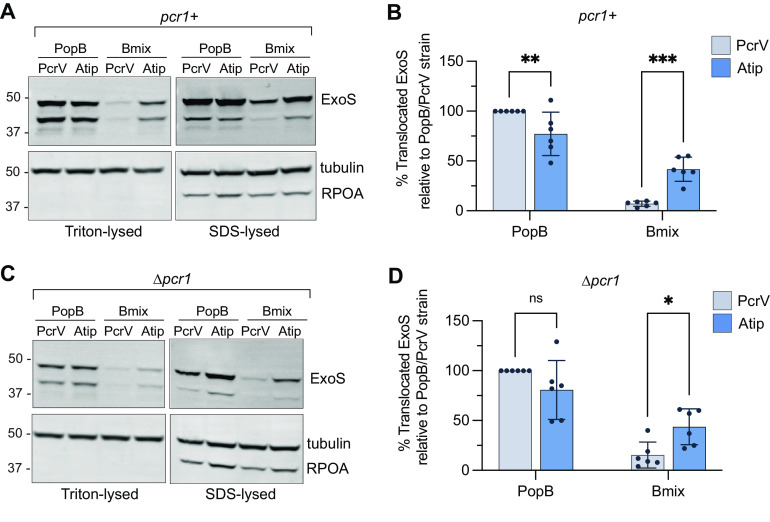
FIG 5.
Deletion of the secretion regulator Pcr1 does not restore translocation for the translocator-mismatched (Bmix-PcrV) strain. A549 cells were infected for 2 h with strain RP3670 [PAO1 ΔexsE exoS(G/A−) pcrV+ ΔpcrH-popBD], strain RP11222 [PAO1 ΔexsE exoS(G/A−) pcrV::Atip ΔpcrH-popBD], strain RP6370 [PAO1 ΔexsE exoS(G/A−) pcrV+ ΔpcrH-popBD Δpcr1], or strain RP11226 [PAO1 ΔexsE exoS(G/A−) pcrV::Atip ΔpcrH-popBD Δpcr1] complemented with popD and the indicated B-translocator, along with both translocator chaperones, pcrH and acrH. The amount of translocated ExoS was assessed by lysing the host cells with either Triton X-100, which lyses the eukaryotic cell membrane but does not lyse the bacteria, or lysing with SDS, which lyses both the eukaryotic cell and peripherally attached bacteria. ExoS was also monitored in supernatant samples and in bacteria collected from the supernatant after infection (shown in Fig. S7). The presence of ExoS, tubulin (host cell cytoplasmic content), and RNA polymerase subunit α (RPOA, bacterial cytoplasmic content) was assessed by Western blotting. (A and C) Western blots of pcr1+ strains (A) and Δpcr1 strains (C). The amount of ExoS translocated into A549 cells is shown by the Triton-lysed samples. The lower-molecular-weight band is attributable to intracellular cleavage of ExoS (59). The positions of molecular-weight size markers (in kilodaltons) are indicated. SDS-lysed samples show the total amount of ExoS both translocated into host cells and remaining in attached bacteria. (B and D) Quantification of ExoS translocation in Triton-lysed samples for pcr1+ strains (B) and Δpcr1 strains (D). The amount of translocated ExoS is normalized to the amount of tubulin in the same strain and to the amount of ExoS in the strain producing PopB/PcrV. n = 6 biological replicates. Error bars show standard deviations. Statistical differences were analyzed with two-way ANOVA and the Sidak multiple-comparison test. Significance thresholds: *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ns, not statistically significant (P > 0.05).