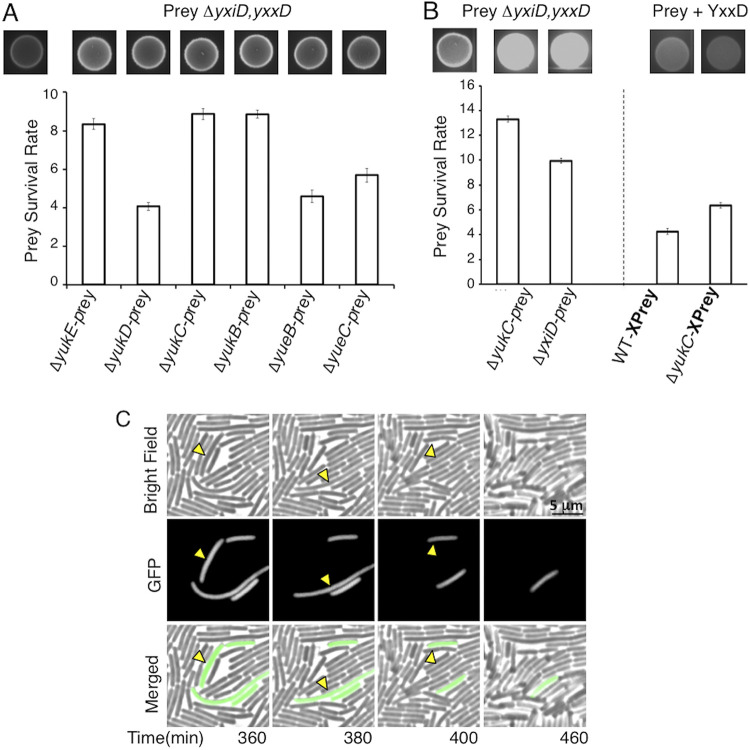
FIG 1.
B. subtilis T7SSb-dependent competition. (A) Competition assay using the fluorescent strain B. subtilis ΔyxiD-yxxD::gfp as the recipient. The wild-type B. subtilis 168 CA strain and the indicated T7SSb mutant derivatives were used as attackers. Top, fluorescence of bacterial spots on agar plate after overnight competition. The spot on the far left resulted from the competition of the prey with the wild=type strain. The other spots were obtained by using attackers with the mutations indicated below. Bottom, prey survival rate plot, indicating that 4 to 9 times more cells survive when the yuk operon genes are deleted in the attacker. The values correspond to the competition spots shown above each plot. (B) Top, fluorescence of bacterial spots on agar plates after overnight competition using a standard prey (left) or a strain complemented for the immunity protein YxxD (right). As a control, the wild-type strain vs the standard prey was used (first spot on the left). Bottom left, prey survival rate plot, showing similar efficiencies when using the ΔyxiC attacker or one depleted of the YxiD toxin (10 to 12 times more cells than in the competing WT/ΔyxiD-yxxD::gfp strain). Bottom right, the YxxD-complemented prey survived the wild-type or the ΔyxiC attacker competition to similar degrees. (C) Effect of YxxD complementation in the recipient strain. In competition assays, the survival of the complemented prey having the yxxD immunity gene reinserted (strain B. subtilis ΔyxxD-yxiD amyE::yxxD) was similar for ΔyukC and wild-type attacker strains. (C) Frames from Movie S1 showing the disappearance of recipient cells from the bright field and fluorescence (GFP) images, as well as their composite images. Examples of cells disappearing in the next frame are highlighted with yellow arrowheads.