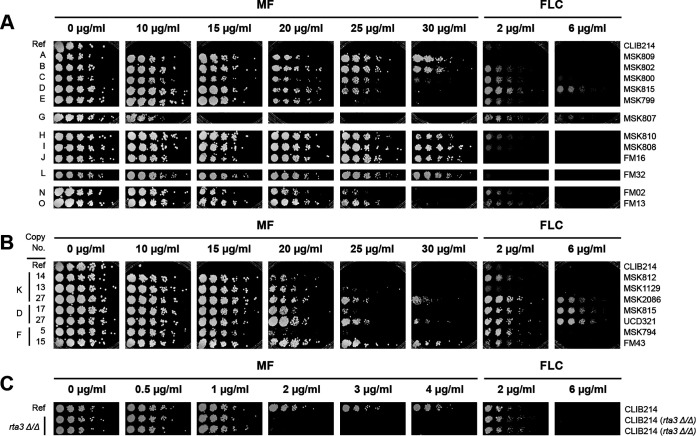
FIG 4.
CNVs of RTA3 correlate with resistance to miltefosine. (A) Multiple RTA3 CNVs are associated with miltefosine resistance. A representative isolate of most CNV patterns (Fig. 2) was grown on YPD with increasing concentrations of miltefosine (MF) or fluconazole (FLC) as shown. Cells were serially diluted (1/5). The CNV pattern is indicated on the left, and the strain name is shown on the right. MF and FLC plates were incubated for 48 h. “Ref” indicates the reference strain, C. parapsilosis CLIB214. (B) Copy number is directly correlated with miltefosine resistance. Isolates with different copy numbers of CNV-K, CNV-D and CNV-F were grown as in (A). The RTA3 copy number of each isolate is shown on the left. (C) Deleting RTA3 reduces resistance to miltefosine. RTA3 was deleted in C. parapsilosis CLIB214 using CRISPR-Cas9. The growth of two biological replicates is shown as in (A), except that the incubations on FLC were for 72 h.