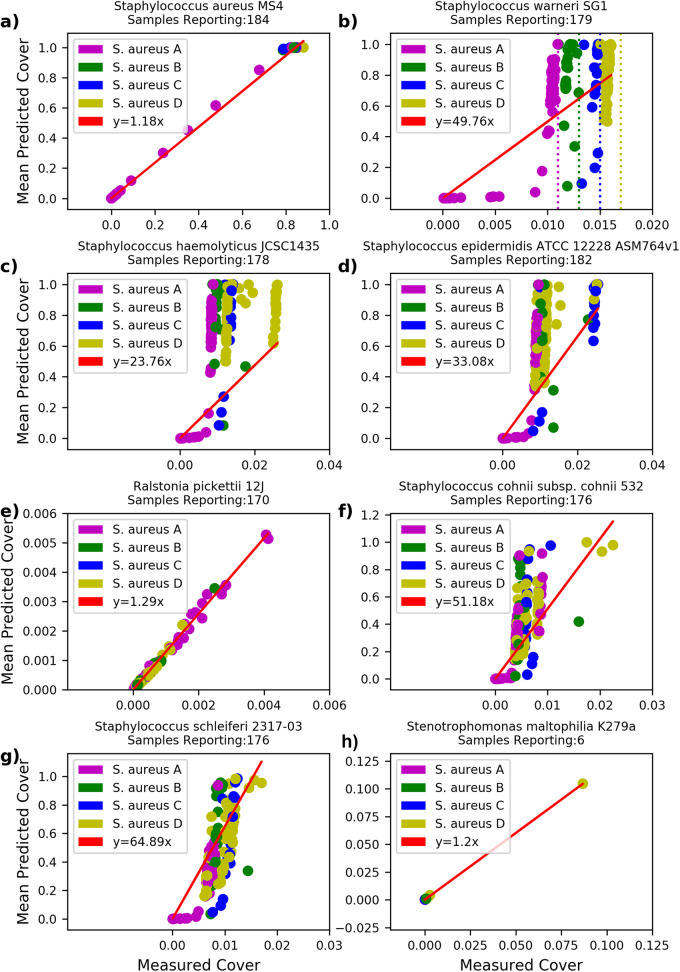
FIG 1.
Modeling genome cover by read count differentiates low abundance contaminants from overlapping references in S. aureus monocultures. Clusters (A) to (D) determined by thresholding of Staphylococcus warneri SG1. Red indicates the line of best fit. A reported slope of 1 with no residual would indicate a perfect model fit. Mean predicted cover calculated using assigned read count and genome length assuming fixed 150 bp read length. As the number of reads increases, measured cover asymptotically approaches the overlap between true sample content and the assigned reference genome.