FIG 3.
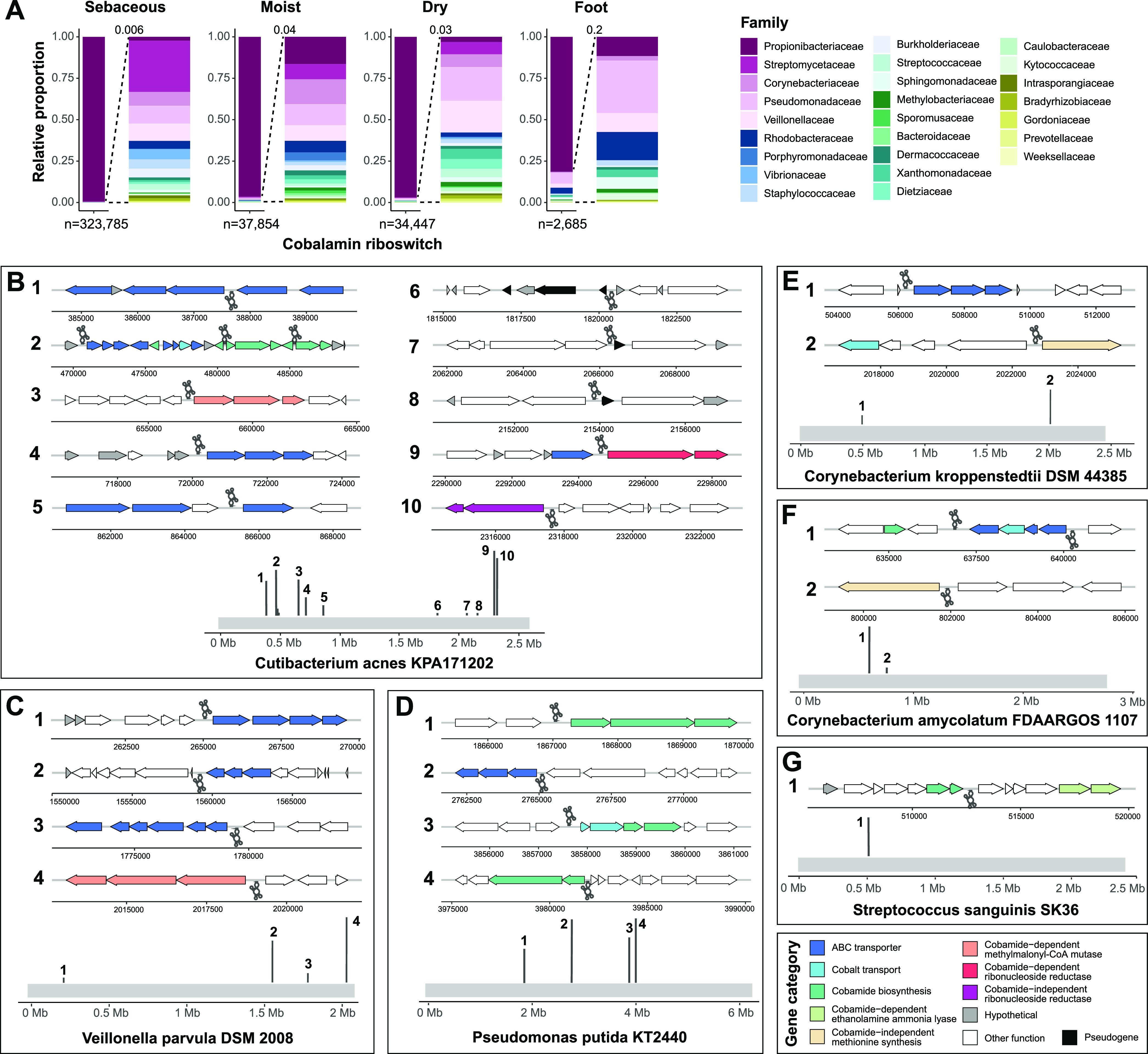
Cobalamin riboswitch regulation varies across skin taxa. (A) The taxonomic abundance of hits for cobalamin riboswitches (Rfam clan CL00101) are shown, with an expanded view of low abundance hits to the right. Total cobalamin riboswitch hits within each microenvironment are indicated. (B to G) Cobalamin riboswitch-containing reads identified from INFERNAL analysis were aligned to Cutibacterium acnes KPA171202 (B), Veillonella parvula DSM 2008 (C), Pseudomonas putida KT2440 (D), Corynebacterium kroppenstedtii DSM 44385 (E), Corynebacterium amycolatum FDAARGOS 1107 (F), and Streptococcus sanguinis SK36 (G) genomes. Dark gray lines along the light gray genome track indicate the position of mapped INFERNAL hits within the genome. Genes upstream and downstream of the riboswitches are colored by their general functional annotation. White (other function) indicates genes not currently known to be associated with cobamides. Gray (hypothetical) indicates a hypothetical protein that has no functional annotation. Right-facing gene arrows and upright dark gray riboswitch icons indicate forward strand orientation, and left-facing gene arrows and inverted riboswitch icons indicate reverse strand orientation. Genomic regions are not to scale.
