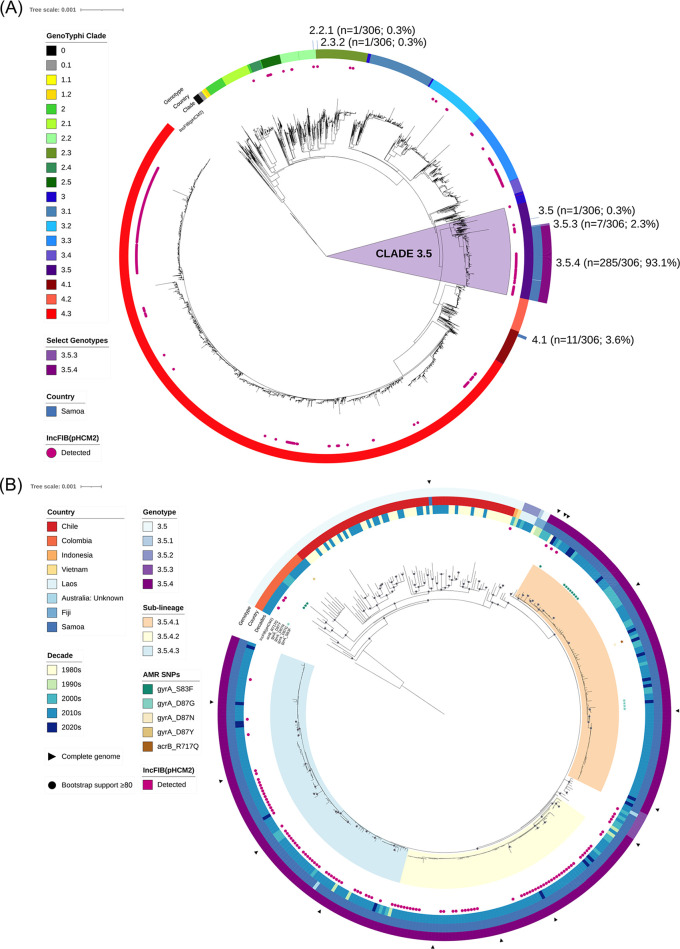
FIG 2.
Phylogenetic analysis of Samoan S. Typhi genomes in a global and dominant clade-specific context. (A) Maximum-likelihood tree of 5,240 S. Typhi genome assemblies using reference strain CT18 (GenBank accession no. AL513382.1) including 306 from Samoa with GenoTyphi clade (ring 1; innermost ring) (9, 74), country (ring 2; Samoa), and genotypes 3.5.4 and 3.5.3 (ring 3) are labeled. The distribution of the IncFIB(pHCM2) replicon detected with PlasmidFinder (82) is indicated with pink shaded circles. Clade 3.5 genomes are highlighted with a light purple background. (B) Maximum-likelihood phylogeny of assembled clade 3.5 genomes (N = 394) using a local Samoan reference strain H12ESR00394-001 (GenBank accession no. LT904890.1). Country of origin, decade of isolation, antimicrobial resistance point mutations, and the detection of the IncFIB(pHCM2) replicon are displayed.