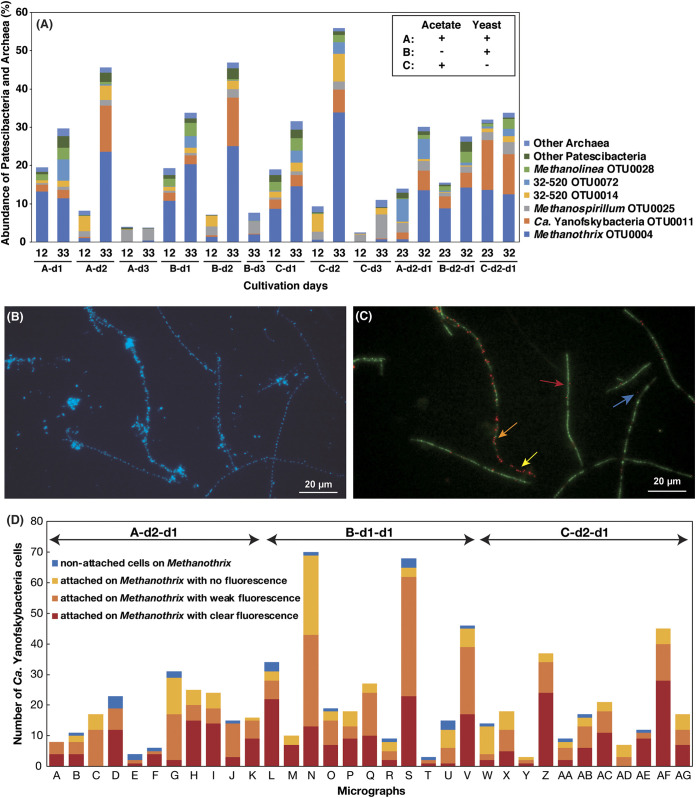
FIG 1.
(A) Relative abundance of predominant Candidatus Patescibacteria and Archaea in the culture systems based on 16S rRNA gene sequence analysis. Micrographs of (B) 4′,6-diamidino-2-phenylindole dihydrochloride staining and (C) fluorescence in situ hybridization (FISH) obtained from culture system B-d1-d1 on day 23 as well as the (D) counted numbers of Ca. Yanofskybacteria cells in 33 micrographs. (C) The microorganisms in the panel were labeled with Ca. Yanofskybacteria-targeting Pac_683-Cy3 probe (red) and Methanothrix-targeting MX825-FITC probe (green). Blue, yellow, orange, and red arrows in (C) indicate nonattached Ca. Yanofskybacteria cells on Methanothrix, attached on Methanothrix with no fluorescence, attached on Methanothrix with weak fluorescence, and attached on Methanothrix with clear fluorescence, respectively, and these colors are consistent with those in the bar diagrams of (D). Culture systems were amended with (A) acetate and yeast extract, (B) yeast extract, and (C) acetate for carbon sources. A-d2-d1, B-d1-d1, and C-d2-d1 were subcultures transferred from A-d2, B-d1, and C-d2, respectively.