Figure 1.
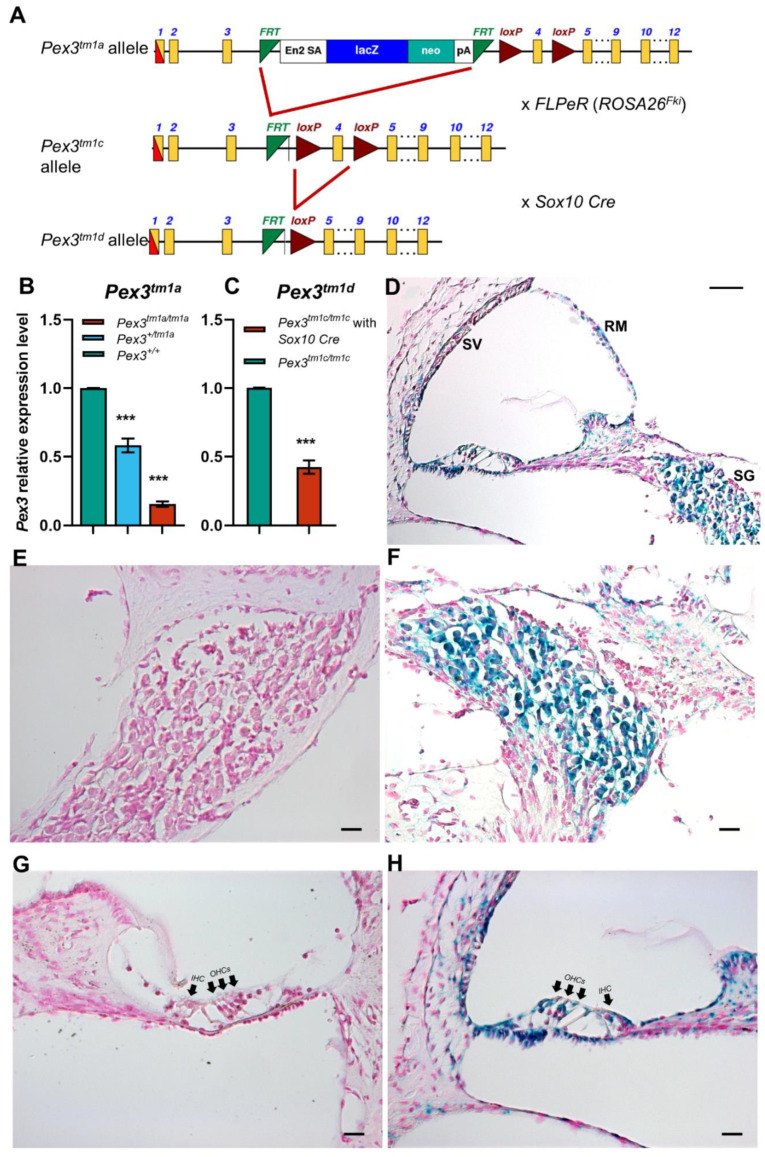
(A) The design of the Pex3tm1a allele is shown at the top. Exposure to Flp recombinase from the FLPeR allele deleted the mutagenic cassette, restoring transcription in the Pex3tm1c mutant. Exposure to Cre recombinase from the Sox10Cre allele deleted exon 4, also leading to a frameshift mutation in the Pex3tm1d allele. Yellow boxes, exons. Green triangles, FRT sites. Red triangles, LoxP sites. The black arrows, in intron 3 of the Pex3 gene, represent the Pex3 forward and reverse primers used for genotyping. The red arrow is the CasR1 primer, used in combination with the Pex3 forward primer to detect the presence of the tm1a allele, and the purple arrows are the primers used to detect the neomycin resistance gene within the tm1a allele. (B) qPCR of brain tissues from 4-week-old mice shows reduced mRNA levels of Pex3 expression in Pex3tm1a homozygotes (16%, red) compared with wildtype littermate controls (green), and heterozygotes have intermediate levels of transcript (58%, blue). The qPCR probe detected sequence downstream of the cassette, from exons 10–11. Data plotted as mean ± standard deviation. ***, p ≤ 0.001, Mann–Whitney rank sum test, n = 7 for homozygotes and wildtypes and n = 6 for heterozygotes. (C) qPCR of cochlear tissues from 4-week-old Pex3tm1d mutant mice shows that Pex3 transcription level was decreased to approximately 42% in the mutants compared with the Pex3tm1c control level. Data plotted as mean ± standard deviation. ***, p = 0.0002. Welsh t-test (normal distribution, but not equal variance). n = 4 for each genotype. (D–H) The reporter gene LacZ was used to reveal the normal expression pattern of Pex3, shown in blue, in the spiral ganglion (D,F) and cell types around the lining of the cochlear duct (D,H) in heterozygous mice aged 2 weeks old. No blue label was detected in wildtype littermates that did not contain the LacZ reporter allele (E,G). IHC, inner hair cells. OHC, outer hair cells. SV, stria vascularis. SG, spiral ganglion. RM, Reissner’s membrane. Scale bars, (D), 50 μm; (E–H), 20 μm. n = 6 for each genotype.