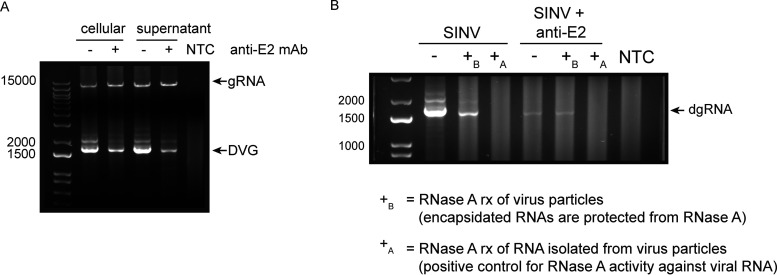
FIG 5.
The nsP1 defective viral genome is packaged and released into viral particles. Differentiated AP-7 cells were infected with SINV (MOI of 10) and treated with medium (mock) or anti-E2 antibody (5 μg/mL) at 4 h after infection. Eighteen hours after infection, RNA from the cells and supernatant fluid was collected. (A) RNA was extracted from the cellular and supernatant samples. RT-PCR analysis for SINV genomic RNA (gRNA) and nsP1 defective viral genome RNA (DVG) was performed. “NTC” represents the no-template control. (B) RNase A protection assay. Culture supernatant was treated with RNase A (10 μg/mL) to digest all nonencapsidated RNAs (+B [before RNA extraction]) or untreated (−). Following RNase treatment, RNA was extracted from the supernatant and reverse transcribed for RT-PCR analysis of nsP1 defective genome RNA (dgRNA). As a positive control for RNase activity, extracted RNA was also treated with RNase A under the same conditions (+A [after RNA extraction]).