FIG 6.
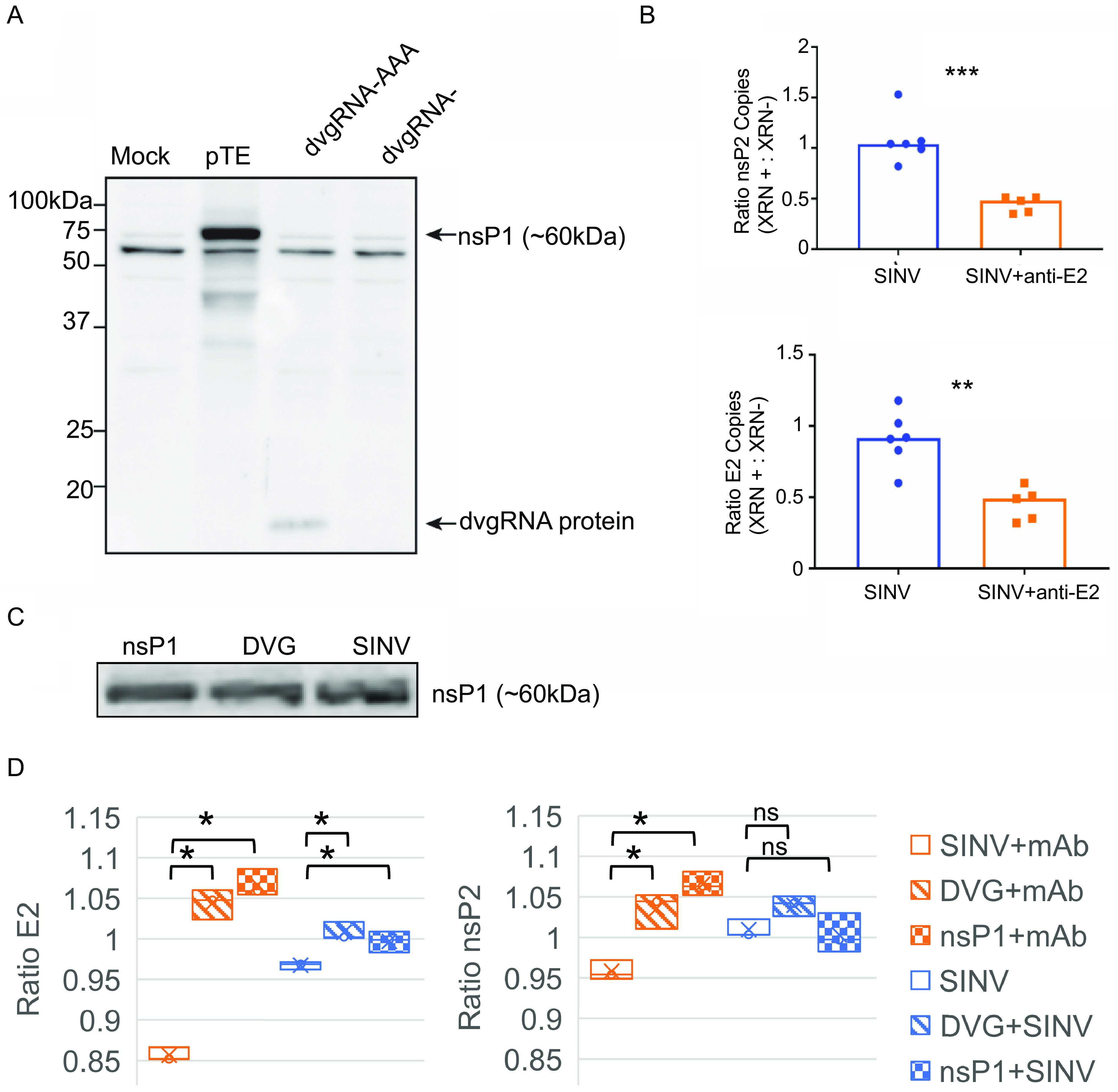
The nsP1 defective viral genome RNA can be translated, and expression is associated with increased SINV RNA capping efficiency. (A) The shorter nsP1 defective viral genome RNA (subDVG) was cloned into the pcDNA3.1 DNA expression vector and in vitro transcribed to RNA (dvgRNA). The RNA was capped and polyadenylated in vitro, purified, and transfected into uninfected undifferentiated AP-7 cells (dvgRNA-AAA). As controls, pTE (complete SINV plasmid) and unpolyadenylated defective viral genome RNA (dvgRNA−) were also transfected. Twenty-four hours after transfection, lysates were collected and assessed by immunoblot probing with a polyclonal antibody against SINV nsP1. (B) Proportion of capped genomes in virus particles released from untreated and antibody-treated SINV-infected dAP-7 cells. Differentiated AP-7 cells were infected with SINV (MOI of 10) and treated with medium (mock) or anti-E2 antibody (5 μg/mL) 4 h after infection. Twenty-four hours after infection, virus particles were purified from the supernatant fluid by ultracentrifugation and viral RNA was extracted and treated with XRN-1 exoribonuclease to degrade noncapped RNAs (XRN +) or left untreated (XRN −). Following treatment, the RNA was reverse transcribed to cDNA and assessed by qRT-PCR for SINV nsP2 and E2 transcripts. The y axis indicates the ratio of copy numbers of nsP2 or E2 transcripts in the XRN-treated versus untreated groups. (C) Immunoblot of nsP1 protein expression 24 h after SINV infection or transfection with pGenLenti vectors expressing nsP1 or nsP1 DVG; (D) proportion of capped genomes in virus particles released from untreated (blue) or antibody-treated (red) cells transiently transfected with lentivirus vector plasmids expressing either nsP1 or nsP1 DVG 24 h prior to infection with SINV. Virions were isolated with Viraffinity reagent, and RNA capping was analyzed using XRN degradation, as described above. Data are presented as mean ± SD and are from three biological replicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
