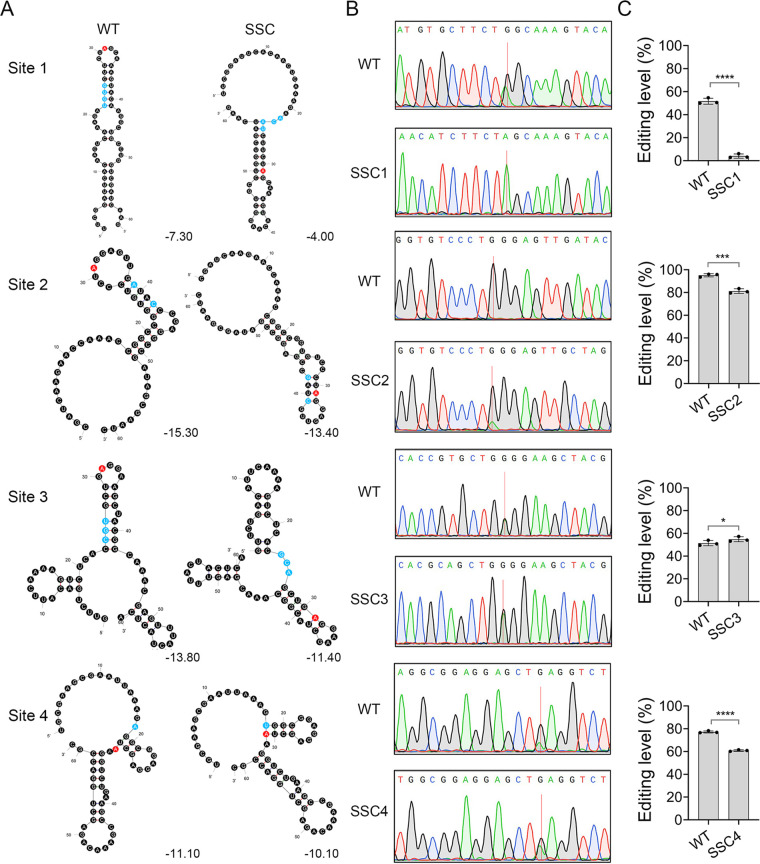
FIG 4.
Effect of secondary structures on RNA editing of FG3G34330. (A) Predicted RNA secondary structures for wild type (WT) and secondary structure change (SSC) sequences of FG3G34330 containing the editing sites together with their upstream and downstream 30 nt sequences. Red, editing sites; Blue, mutation sites for secondary structure changes. The minimum free energy (MFE) value of each predicated RNA structure is indicated. (B) Sequencing traces of the 4 editing sites and their flanking sequences amplified from RNA isolated from perithecia of WT and marked mutants. Red lines mark the editing sites with mixed peaks of A and G although only the dominating peak is shown in the sequence on the top. (C) Means and standard deviations of the editing levels were estimated from three biological replicates (n = 3). Significant differences for pairwise comparison are based on t test (*, P < 0.05; ***, P < 0.001; ****, P < 0.0001).