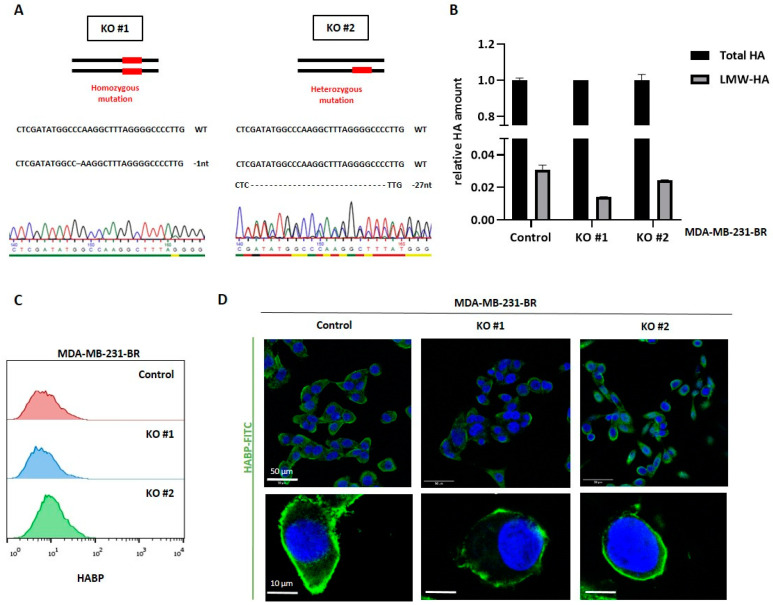
Figure 3.
Generation of HYAL1 knockout (KO) in MDA-MB-231-BR and characterization regarding their HA metabolism. (A) Two types of HYLA1 mutations generated by CRISPR/Cas9 in MDA-MB-231-BR cells were identified after Sanger sequencing in the corresponding sequencing chromatogram. Mutation features shown here include a homozygous single-base deletion in clone KO #1 (indicated as -1nt) and a larger deletion in only one allele of clone KO #2 (-27nt). (B) Bar graph displaying representative HA-ELISA results on secreted HA levels (one representative experiment with duplicates out of three experiments is shown (n = 3)). The amount of LMW-HA is presented as a ratio of total HA (set as 1). (C) Multilayer histogram showing the quantified amount of surface HA measured via flow cytometry, using biotinylated HA binding protein and FITC-conjugated Streptavidin. One representative experiment is shown (n = 3). (D) Immunofluorescence staining of nuclei (DAPI, blue) and HA (FITC, green) using biotinylated HABP and FITC-conjugated Streptavidin. Scale bars represent 50 µm in the upper panel and 10 µm in the lower panel. Statistical significance was determined using unpaired two-tailed Student’s t-tests. The assumption of homogeneity of variance was tested using Levene’s Test of Equality of Variances (p > 0.05). Values are means +/− s.d.