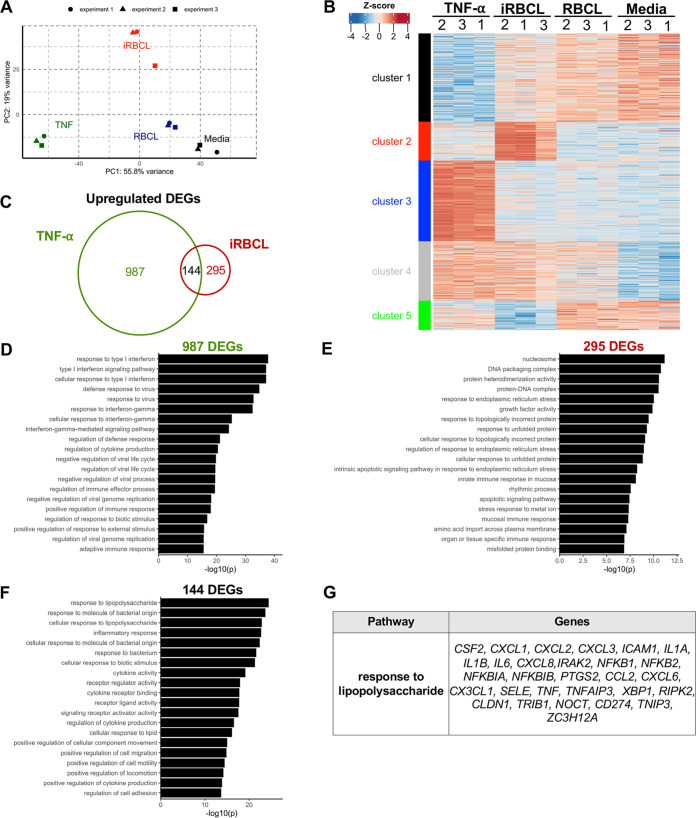
FIG 5.
TNF-α and P. falciparum-iRBC lysates induce distinct transcriptional profiles, with overlap in genes associated with inflammation. HBMEC monolayers were incubated with medium, TNF-α (1 ng/mL), RBCLs (8 × 106/cm2), or P. falciparum-iRBCLs (8 × 106/cm2) for 6 h prior to RNA extraction and sequencing. (A) Principal-component analysis of 2,373 differentially expressed genes (DEGs). Sample points correspond to independent experiments. (B) Heat map of the 2,373 DEGs with K-means clustering to distinguish uniquely regulated sets of genes. Experimental samples for all conditions are shown and denoted with 1, 2, or 3. (C) Overlap analysis of TNF-α versus medium (TNF-α) and iRBCL versus RBCL (iRBCL) DEGs. Cutoffs of DEGs were set to log2 fold change of >1 or <−1 with an adjusted P value of <0.05. (D to F) Gene Ontology (GO) term enrichment analysis of DEGs from panel C. (D to F) Top 20 pathways mapped by upregulated DEGs (D) unique to TNF-α, (E) unique to iRBCLs, and (F) shared by both. (G) Gene list of the top pathway from panel F.