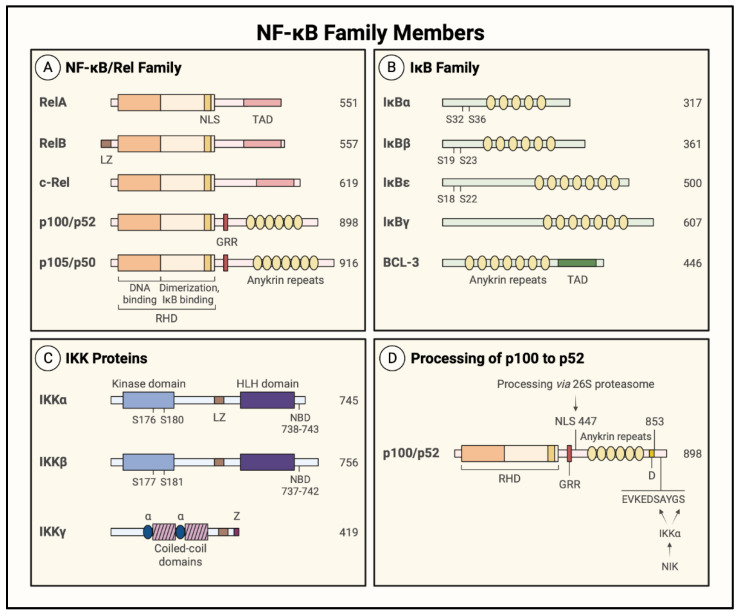
Figure 5.
Structure of NF-κB family members. The number of amino acids in each protein is listed on the right. (A) The five members of the NF-κB family contain a Rel homology domain (RHD) at their N-terminus. The RHD is comprised of a DNA binding domain, a dimerization/IκB binding domain, and a nuclear localization sequence (NLS). At their C-terminus, the catalytically active RelA, RelB, and c-Rel proteins contain transactivation domains (TADs), whereas p105 and p100 contain a glycine-rich region (GRR) and ankyrin repeats (ovals). The p105 and p100 proteins are catalytically inactive due to the presence of their ankyrin repeats, and the GRR facilitates the cotranslational processing of p105 to p50 and the post-translational processing of p100 to p52. (B) The ankyrin repeats (ovals) in IκB proteins interact with the RHD of NF-κB family members and mask its NLS, and serine phosphorylation at the indicated sites promotes their proteasomal degradation. BCL-3 also contains a TAD at its C-terminus, which confers transcriptional activity to p50/BCL-3 and p52/BCL-3 complexes. (C) The catalytically active IKKα and IKKβ proteins contain kinase and helix–loop–helix (HLH) domains, and their activity is stimulated by phosphorylation of the kinase domain at the indicated serines. The regulatory protein, IKKγ (or NEMO), contains a zinc finger (Z) domain, two coiled-coil domains, and two a-helical (a) domains, which mediate its association with the NEMO binding domain (NBD) at the C-terminus of IKKα and IKKβ. All three members also contain a leucine zipper (LZ) motif required for protein–protein interaction. (D) NF-κB-inducing kinase (NIK) activates IKKα, which phosphorylates the p100 C-terminus to stimulate the post-translational processing of p100 to p52. The p100 protein is cleaved at amino acid 447, and the C-terminus is degraded by the 26S proteasome. The GRR serves as a signal to prevent the proteasome from degrading the N-terminal portion of the protein.