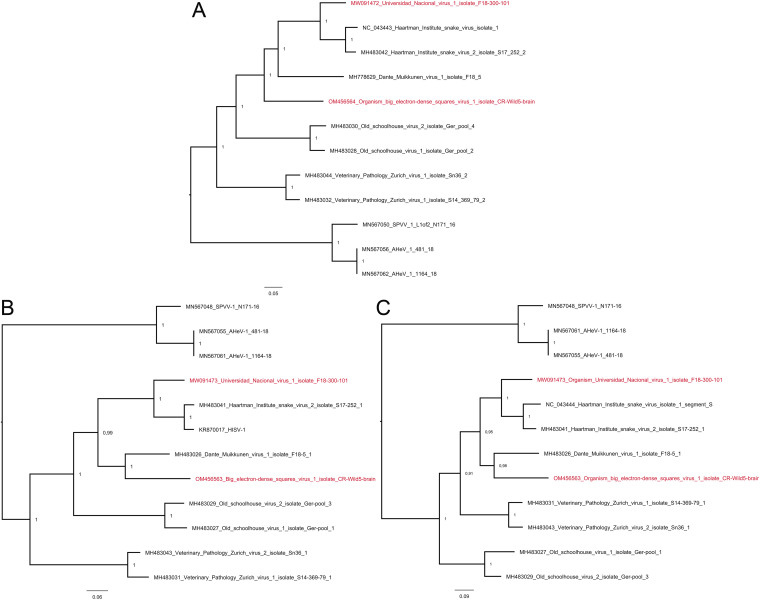
FIG 6.
Phylogenetic analysis of hartmaniviruses identified in the study. The maximum clade credibility trees were inferred using the Bayesian MCMC method with Cprev Blosum and WAG amino acid substitution models for RdRp, GPC, and NP, respectively. (A) A phylogenetic tree based on the RdRp amino acid sequences of the viruses identified in this study and those available in GenBank. (B) Phylogenetic tree based on the NP amino acid sequences of the viruses identified in this study and those available in GenBank. (C) A phylogenetic tree based on the GPC amino acid sequences of the viruses identified in this study and those available in GenBank.