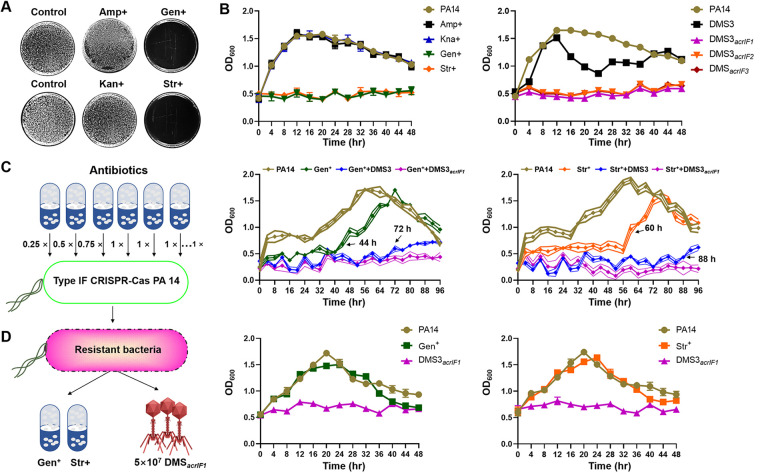
FIG 6.
DMS3acrIF1 ameliorates P. aeruginosa antibiotic resistance. (A) PA14 (OD600 = 0.5) was evenly spread on a plate containing different antibiotics (Amp+: 100 μg/mL, Kan+: 100 μg/mL, Gen+: 100 μg/mL, Str+: 100 μg/mL), and antibiotic resistance was tested by observing the growth of bacteria on the plate. (B) PA14 (5 × 108 CFU/mL) was cultured in lysogeny broth (LB) containing different antibiotics (Amp+: 100 μg/mL, Kan+: 100 μg/mL, Gen+: 100 μg/mL, Str+: 100 μg/mL) or treated with 1 × 108 PFU/mL EATPs/DMS3 (MOI = 0.2). The growth kinetics of PA14 were continuously monitored at 4 h intervals up to 48 h at 37°C with constant shaking. (C) The left panel is a schematic diagram of the antibiotic resistance experiment. PA14 (PA14: 5 × 108 CFU/mL) was treated with gradient concentrations of antibiotic (Gen+ or Str+), gradient concentrations of antibiotic and DMS3 (MOI = 0.02) (Gen+/Str+ + DMS3) or Gen+/Str+ + DMS3acrIF1. The growth kinetics of PA14 were measured. (D) PA14 that acquired antibiotic resistance were picked out and grown to the mid-logarithmic phase (OD600 = 0.4 to 0.6) in LB at 37°C with 220 rpm shaking. The growth kinetics of the acquired antibiotic resistance of PA14 were measured after treatment with DMS3acrIF1 (MOI = 0.2) or with 100 μg/mL Gen+/Str+ antibiotics. Data were presented as mean ± standard error of the mean (SEM), determined from biological triplicates and analyzed via a one-way ANOVA compared against a control group (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).