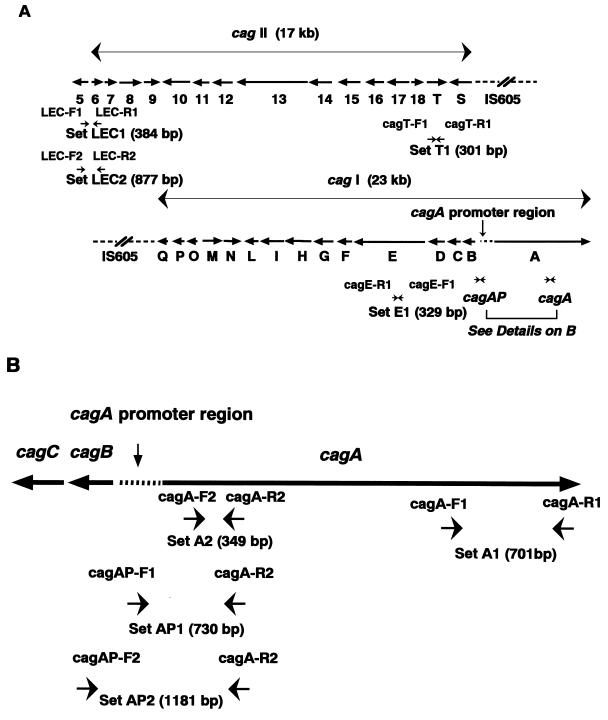
FIG. 1.
Structure of cag pathogenicity island and locations of PCR primers. (A) Locations of PCR primers for cagE, cagT, and the LEC. Two sets of primers were used to detect the cagA gene: primer set A1 (cagA-F1 and cagA-R1) and set A2 (cagA-F2 and cagA-R2). For detection of the promoter region of cagA, primer sets AP1 (cagAP-F1 and cagA-R2) and AP2 (cagAP-F2 and cagA-R2) were used, and for the left end of cagII, primer sets LEC1 (LEC-F1 and LEC-R1) and LEC2 (LEC-F2 and LEC-R2) were used. The expected lengths of PCR products using each primer set are shown. The sequence and location of each primer are shown in Table 1. (B) Locations of PCR primers for the cagA gene and its promoter region. The expected lengths of PCR products using each primer set are also shown.