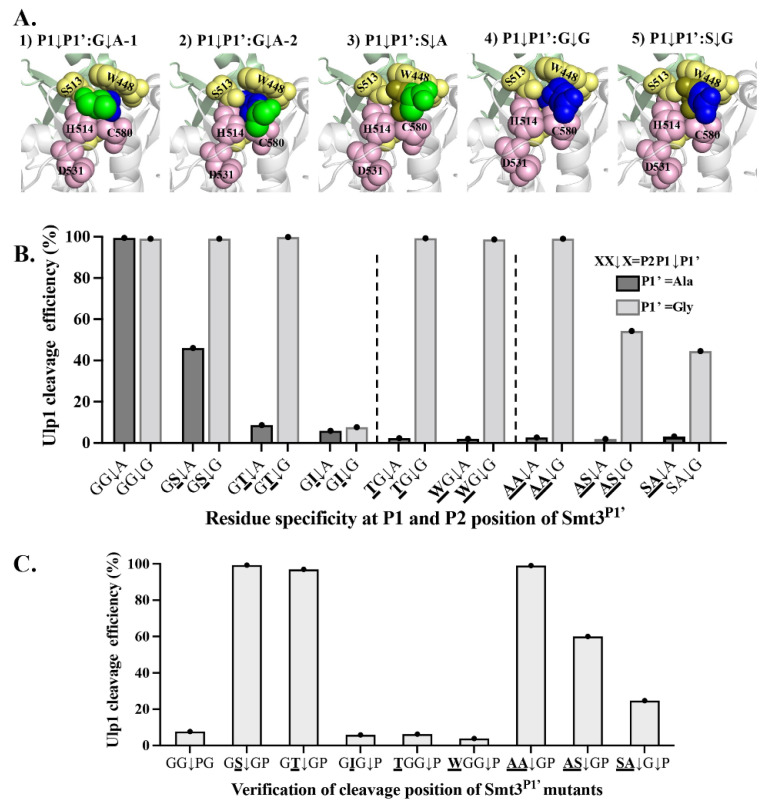
Figure 5.
P1’ position Gly expands residue inclusivity of Ulp1 against P1 and P2 position of Smt3. (A) The substrate pocket structure of Ulp1–Smt3 and Ulp1–Smt3P1P1’ complex at P1 and P1’ position. Blue is P1 position Gly; green is P1’ position Ala; and brown is P1 position Ser. (1) The first conformation of Smt3–GG↓A substrate’s target peptide bond between P1–P1’ position (GG↓A) requires enough flexibility to approach nucleophilic residue C580 for cleavage. (2) The second conformation of Smt3–GG↓A substrate’s P1’ position Ala side chain extension may cause obstruction in Ulp1 catalytic residues H514 and C580 ability to affect cleavage efficiency. (3) P1 position amino acids with longer side chains, like Ser, would limit flexibility of C-terminal tail in the tunnel, which may prevent P1’ position Ala from rotating to remove the steric hindrance in Ulp1 catalytic residues, and the P1–P1’ peptide bond was not close to catalytic residue C580. Therefore, Ulp1 cleaving efficiency was affected. (4) P1’ position Gly has high flexibility and does not form hindrance to Ulp1 catalytic residues H514, C580 and D531. (5) P1 or P2 position residues, like Ser, do not require a huge free space to guarantee high flexibility of P1’ position Gly. (B) The residue specificity of Ulp1 on Smt3P when P1’ position was Ala or Gly. P2P1↓P1’ position residue in wild-type Smt3 is GG↓A. (C) Verification of cleavage position in Smt3P substrates when P1’ was Gly. P2P1∣P1’P2’ position residue in wild-type Smt3 is GG↓AT. Cleavage efficiency of Ulp1 against corresponding substrates is presented as average cleavage efficiency of three independent experiments.