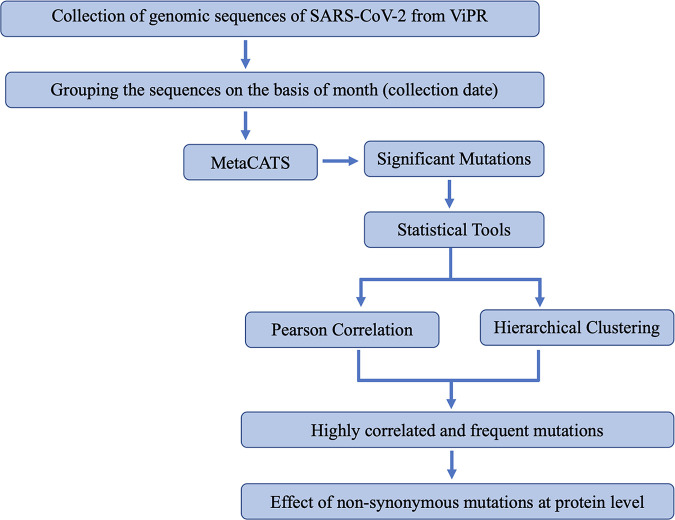
FIG 1.
Methodology used in this study. The SARS-CoV-2 complete genome sequences were obtained from the ViPR and grouped by month based on the collection date. The meta-analysis was then performed in a pairwise manner, i.e., comparing January 2020 to each month through March 2021, to identify the highly significant mutations arising among the genomic sequences of SARS-CoV-2. Once the significant mutations were identified, we used Pearson correlation- and hierarchical clustering-based approaches to identify correlations and clusters among the highly significant mutations. The effect of these mutations on the wild-type protein was then studied using several computational tools, including PredictSNP, ENCoM ΔΔSVib, DynaMut, ENCoM ΔΔG, mCSM ΔΔG, DUET ΔΔG, and SAAFEC-SEQ.