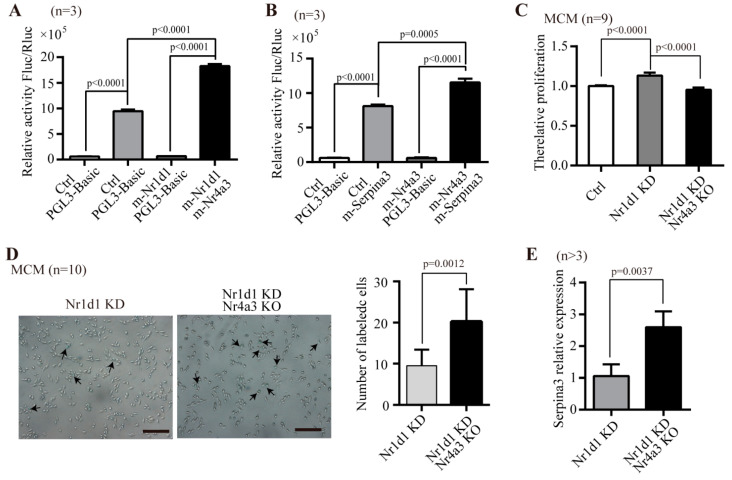
Figure 7.
Regulatory mechanism of Nr1d1. (A,B) effect of Nr4a3 and Serpina3 on transcription of Nr1d1 assessed by a Dual-Luciferase reporter system. MCM cells were transfected with Nr4a3 luciferase reporter plasmid for 24 h, and incubated with or without the Nr1d1 plasmid (A) or MCM cells were transfected with Serpina3 luciferase reporter plasmid for 24 h, and incubated with or without the Nr4a3 plasmid (B). Cells were lysed with lysis buffer, and the lysates were subjected to the luciferase activity assay, data were collected from three independent experiments (n = 3), two-way ANOVA was applied for statistical analysis, and p < 0.05 was considered significant. (C–E) knockdown of Nr4a3 in MCM cells by CRISPR/Cas9 technique, followed by downregulation of Nr1d1 using Nr1d1-shRNA lentivirus transduction; (C) the effect of Nr4a3 knockout on cell proliferation in the MCM cells with Nr1d1 knockdown. The data were from three independent experiments, and each experiment had three replicates (n = 9). One-way ANOVA was used for statistical analysis and p < 0.05 was considered significant; (D) the effect of Nr4a3 knockout on cellular senescence (beta-galactosidase staining) in MCM cells with Nr1d1 knockdown. The image on the left panel is a representative field of view, scale bars: 50 μm. The bar graph on the right panel shows the number of cells that were stained blue in each random field of view. Each experiment was repeated three times, and 10 random fields of view were counted (n = 10). Two-way ANOVA was used for statistical analysis, and p < 0.05 was considered significant. (E) The Serpina3 mRNA expression levels were detected by real-time PCR on Nr1d1 knockdown MCM cells versus on Nr1d1 knockdown and Nr4a3 knockdown MCM cells (n = 3 vs. 5); a two-tail unpaired t-test was used as an analysis method, p < 0.05 was considered statistically significant. All data in bar graph presented as mean ± SD. Ctrl: control, KD: knockdown, KO: knockout.