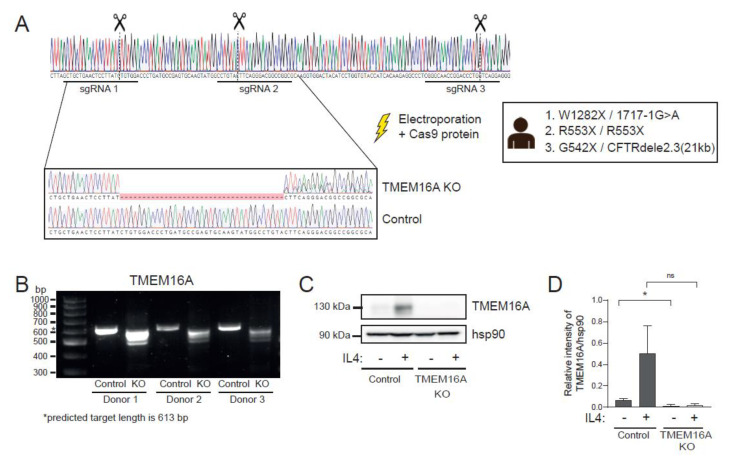
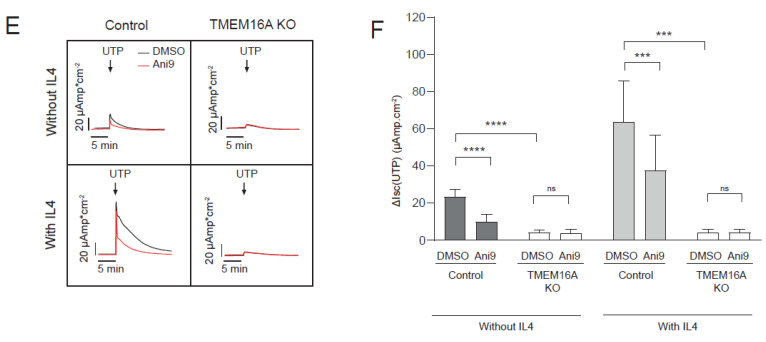
Figure 5.
Generation and validation of TMEM16A KO nasal epithelial cells. CRISPR-Cas9 based gene editing was used to generate a TMEM16A KO in CFTR-null nasal cells. (A) Graphical overview showing binding sites of three sgRNA molecules and Sanger sequencing traces after electroporation; (B) DNA gel showing the PCR-amplified products of the targeted TMEM16A locus for KO and control samples of 3 CFTR-null donors (G542X/CFTRdele2.3 (21 kb), W1282X/1717-1G>A, R553X/R553X). Predicted length of the PCR product was 613 bp; (C) representative Western blot for TMEM16A protein of ALI-differentiated TMEM16A KO and control cells. To increase TMEM16A expression, some cells were treated with IL-4 for 48 h; (D) quantified band intensity of TMEM16A protein in Western blots (n = 3 independent donors); (E) functional validation of ALI-differentiated TMEM16A KO cells with Ussing chamber measurements. TMEM16A activity was determined based on Ani9-sensitive (1 µM) UTP-induced (100 µM) currents. Representative traces are shown of one donor and (F) UTP-induced currents were quantified for all donors, with and without Ani9-treatment (n = 3 independent donors). All cells were treated with amiloride and indicated cells were treated with IL-4 for 48 h. Analysis of differences was performed using paired t-tests (D) or a 2-way ANOVA with Tukey post hoc test (F). ns = non-significant, * p < 0.05, *** p < 0.001, **** p < 0.0001.