Figure 3.
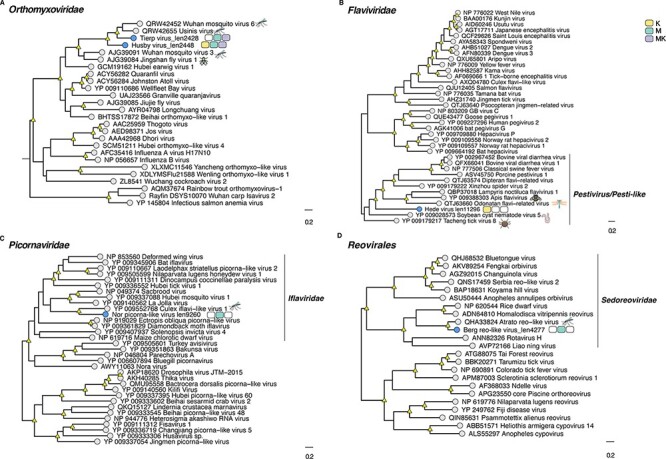
Phylogenetic relationships among the viruses found in this study and reference sequences within the RNA virus families (A) Flaviviridae, (B) Orthomyxoviridae, (C) Picornavirales, and (D) Sedoreoviridae. Phylogenetic trees were estimated using the VT + F + I + G4 (Flaviviridae, Picornavirales, and Sedoreoviridae) and Q.pfam + F + I+ Γ4 (Orthomyxoviridae) substitution models. Novel viruses are indicated by blue tip points, and hosts are represented with three-pack bars corresponding to mite (K; yellow), mite-free mosquito (M; green), and mite-detached mosquito samples (MK; purple). Trees are based on the amino acid sequences of the putative RdRp. Nodal support values ≥80 per cent SH-aLRT and ≥95 per cent UFboot are denoted with yellow triangles at nodes. Scale bars indicate the number of amino acid substitutions per site and the trees are mid-point rooted for clarity only. Host species information (animal icons) is shown for the closest relatives of the novel viruses.
