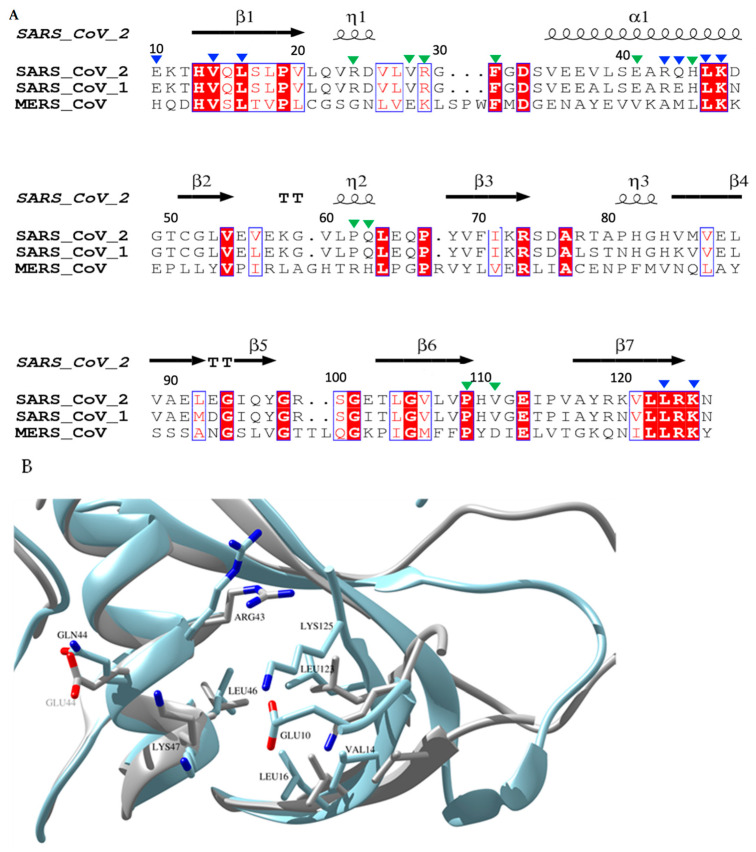
Figure 5.
Sequence and structural alignment of the N-terminal domain of three medically relevant coronaviruses SARS-CoV-2, SARS-CoV-1 and MERS nsp1s. (A) Structural protein sequence alignment of SARS-CoV-1, SARS-CoV-2 and MERS. Key residues in binding pockets I and II are indicated by blue and green triangles, respectively. The sequences were aligned using CLUSTAL W multiple alignment [43] and the secondary structure elements were extracted using ESPript 3.0 [44]. (B) Overlay of SARS-CoV-1 (coloured in light grey) and SARS-CoV-2 (coloured in light blue) highlighting the structural differences around binding pocket I (RMSD of 0.748 Å). Pairwise key residues of the two nsp1s in pocket I interacting with fragment hits are shown in sticks. Glu10 is absent in the NMR structure of SARS-CoV-1 nsp1 (PDB entry 2HSX).