Abstract
Gram-negative bacteria were reported as a significant cause of infections in both community and nosocomial settings. Considered as one of the greatest threats to public health, the spread of bacteria drug resistance and the lack of effective alternative treatment options remains problematic. Herein, we report a promising strategy to combat Gram-negative resistant strains consisting of the combination of a macrolide antibiotic with a polyaminoisoprenyl adjuvant derivative leading to a significant decrease of antibiotic resistance.
Keywords: polyaminoisoprenyl derivatives, antibiotic adjuvants, polyamines, Gram-negative bacterial strains, macrolide antibiotics, outer membrane
1. Introduction
The discovery of beta-lactam antibiotics in 1928 by Fleming was revolutionary and has saved countless lives from severe infectious diseases caused by bacterial strains [1,2,3]. Nevertheless, the excessive use of these drugs in many fields of medicine has contributed to the emergence of bacterial resistance and leading to limited treatment options [4,5]. The fast rate of spread of multidrug-resistant Gram-negative bacteria remains an important concern due to their intrinsic resistance and their ability to rapidly develop new mechanisms of resistance [6,7]. In this context, Enterobacteriaceae, such as Klebsiella pneumoniae and Escherichia coli, as well as Pseudomonas aeruginosa and Acinetobacter spp., have been identified as responsible for most multidrug-resistant bacterial infections [8,9,10]. However, antibiotic resistance represents a natural phenomenon that cannot be stopped and innovative approaches to restore antibiotic failure are desperately needed. In this context, it has been previously demonstrated that combined antibiotic therapy using macrolide antibiotics such as erythromycin, clarithromycin, and azithromycin could present high anti-biofilm activity both in vitro and in vivo [11]. Another approach currently under investigation is the design and synthesis of new classes of adjuvants able to restore the activity of inefficient antibiotics [12].
Gram-negative bacteria are intrinsically resistant to many compounds due to their outer membrane (OM) composition. Thus, it is well known that their cell envelope is a complex multilayered structure where bacteria are surrounded by a thin peptidoglycan cell wall, which itself is surrounded by an outer membrane containing lipopolysaccharide (LPS). In the complex OM architecture lipid A, the hydrophobic group of lipopolysaccharide covers the surface of most Gram-negative bacteria, playing an essential role by anchoring the lipopolysaccharide in the membrane [13]. Furthermore, the spatial organization of LPS molecules is stabilized by Mg2+ and Ca2+ divalent cations, resulting in a barrier that is difficult to penetrate by numerous classes of antibiotics [14,15,16,17,18,19]. Thus, this OM obstacle needs to be circumvented since it has proven to be especially problematic to modern target-based antibacterial drug design. OM perturbation can be achieved in vitro through a variety of genetic means [17,20,21]. However, the effects of such genetic perturbation are mostly permanent [22,23]. On the other hand, well-known compounds such as EDTA increase the permeability of the OM by chelating the divalent cations stabilizing LPS [24,25]. Other chemical compounds, most notably polymyxin derivatives, can bind the lipid A displacing the divalent cations and, thus, disrupting the OM integrity [26,27,28,29,30]. Nevertheless, despite being good OM permeabilizers, their inner membrane (IM) activity makes these compounds mainly nonspecific [31].
For more than six decades, the clinical use of macrolides increased gradually since their discovery [32]. Known for their mechanism of action of inhibiting protein synthesis by targeting the bacterial 50S ribosomal subunit, almost all these drugs present similar antibacterial profiles and are mainly active against Gram-positive bacteria [33]. This family of antimicrobials has also been used against specific and limited species of Gram-negative bacteria such as azithromycin, commonly used against Enterobacteriaceae infections [34].
To discover new compounds capable of perturbing the Gram-negative OM, our group has recently developed the design and use of polyaminoisoprenyl derivatives as antibiotic enhancers exhibiting a strong effect on the level of tetracycline antibiotics susceptibility against resistant P. aeruginosa bacterial strains [35,36,37,38]. In the continuing course of our studies, we report herein a promising strategy to combat Gram-negative resistant strains consisting of the combination of a macrolide antibiotic with polyaminoisoprenyl adjuvant derivatives leading to a significant decrease of antibiotic resistance and increase of Gram-negative strains susceptibility towards macrolides. We have also investigated the efficiency of such an approach against the most common bacterial strains and established a close resistance profile–activity relationship.
2. Results
2.1. Synthesis of Polyaminoisoprenyl Derivatives 3–6
The synthesis of polyaminoisoprenyl derivatives 3–6 utilized an optimized direct nucleophilic substitution of the appropriate polyamine on farnesyl chloride 1 and neryl chloride 2 performed in THF at room temperature for 12 h (Table 1). Performing this reaction for 24 h led to the formation of a higher proportion of by-products. Under the 12 h reaction conditions, the expected products were obtained as pure isomers in yields ranging from 49 to 72%, respectively. Cytotoxicity was evaluated against Chinese hamster ovary (CHO) cells, with all compounds presenting IC50 ranging from 126 to 150 µM, suggesting that they were minimally toxic.
Table 1.
Polyaminoisoprenyl derivatives 3–6 synthesis from farnesyl or geranyl chloride 1–2.
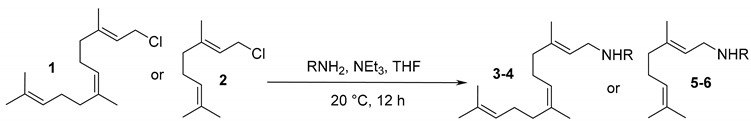
| RNH2 | Cpd-(Isolated Yield (%)) | IC50 (µM) CHO | ||
|---|---|---|---|---|
| 3–4 | 5–6 | |||
|
3 (72) | 5 (64) | 142.79 | >150 |
|
4 (49) | 6 (63) | >150 | 126.82 |
2.2. Antimicrobial Activity of Polyaminoisoprenyl Derivatives 3–6 against Gram-Negative Bacteria
Table 2 summarizes the MICs obtained for the polyaminoisoprenyl derivatives 3–6 against Gram-negative strains. Compounds 3–6 demonstrated a similar behavior with MICs ranging from 50 to greater than 200 µM, whereas compound 3 presented MICs from 12.5 to 100 µM depending on the considered bacterial strains.
Table 2.
Minimum Inhibitory Concentrations of polyaminoisoprenyl derivatives 3–6 against various Gram-negative bacterial strains.
| MIC (µM) (µg/mL) | ||||
|---|---|---|---|---|
| Strains | Cpd 3 | Cpd 4 | Cpd 5 | Cpd 6 |
| P. aeruginosa PA01 | 25 (10) | 200 (78) | >400 (>135) | >400 (>130) |
| E. coli ATCC 25922 | 50 (20) | 200 (78) | >200 (>67) | >200 (>65) |
| K. pneumoniae ATCC 13883 | 50 (20) | 200 (78) | >200 (>67) | >200 (>65) |
| C. koseri IP8294 | 50 (20) | 200 (78) | >200 (>67) | >200 (>65) |
| E. cloacae DSM 129 | 50 (20) | 200 (78) | >200 (>67) | >200 (>65) |
| K. aerogenes ATCC 13048 | 100 (40) | >400 (>156) | >400 (>135) | >400 (>130) |
| K. aerogenes 289 | 100 (40) | >400 (>156) | >400 (>135) | >400 (>130) |
| AG100A_pUC18 | 12.5 (5) | 50 (19) | 100 (33) | >200 (>65) |
2.3. MICs of the Different Macrolides Tested against Various Gram-Negative Bacteria
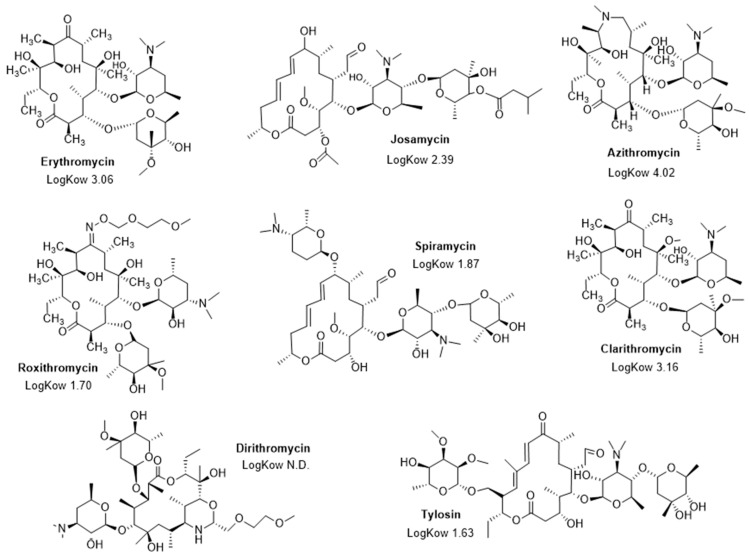
The MICs of different macrolides (erythromycin, josamycin, roxithromycin, azithromycin, spiramycin, clarithromycin, dirithromycin, and tylosin) (Figure 1) were determined against all the Gram-negative bacterial strains, with MIC’s ranging from 2 µg/mL to greater than 1024 µg/mL (Table 3). In this context azithromycin appeared as the most effective antibiotic against all the selected bacteria with MICs varying from 2 to 128 µg/mL, whereas all the other tested macrolides led to higher MICs ranging from 128 to 1024 µg/mL.
Figure 1.
Structure of the macrolides used in this study and their associated LogKow values.
Table 3.
Minimum Inhibitory Concentrations of macrolides against various Gram-negative bacteria.
| MIC (µg/mL) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Strains | Erythro | Josa | Azithro | Roxithro | Spira | Clarithro | Dirithro | Tylo |
| PA01 | 512 | >1024 | 128 | >1024 | >1024 | 512 | 1024 | >1024 |
| E. coli ATCC 25922 | 128 | 1024 | 8 | 512 | 512 | 128 | 64 | 512 |
| K. pneumoniae ATCC 13883 | 128 | 1024 | 16 | 512 | 512 | 128 | 64 | 1024 |
| C. koseri IP8294 | 256 | 1024 | 16 | 1024 | 1024 | 128 | 32 | 1024 |
| E. cloacae DSM 129 | >1024 | >1024 | 64 | >1024 | >1024 | 1024 | 128 | >1024 |
| K. aerogenes ATCC 13048 | 512 | 512 | 64 | 512 | 1024 | 256 | 64 | 1024 |
| K. aerogenes 289 | >1024 | >1024 | 64 | >1024 | 1024 | >1024 | 1024 | >1024 |
| AG100A_pUC18 | 8 | 32 | 2 | 32 | 128 | 64 | 16 | 256 |
Erythro, Erythromycin; Josa, Josamycin; Azithro, Azithromycin; Roxithro, Roxithromycin; Spira, Spiramycin; Clarithro, Clarithromycin; Dirithro, Dirithromycin; and Tylo, Tylosin.
2.4. Restoration of Macrolides Activity against Various Gram-Negative Bacteria in Combination with Derivatives 3–6
MICs of the macrolides in combination with the polyaminoisoprenyl derivatives were determined to evaluate the antibiotic enhancing activities of 3–6 toward numerous Gram-negative bacterial strains (Table 4). Compound 3 used at a 10 µM concentration increased the susceptibility of all the bacterial strains with respect to all the tested antibiotics by improving their antimicrobial activities. It is noteworthy that compound 4 led to some enhancement but in a less efficient manner than compound 3. Interestingly, under the same experimental conditions, the parent geranyl derivatives 5 and 6 demonstrated only weak ability to restore the activity of the macrolides toward Gram-negative bacteria.
Table 4.
Restoration of macrolides activity (MIC values in µg/mL) against various Gram-negative bacteria in the presence of a 10 μM concentration of derivatives 3–6.
| Antibiotic | Cpd | PA01 | E.c. a | K.p. b | C.k. c | E.cl. d | K.a. e | Ea289 f | E.c. g |
|---|---|---|---|---|---|---|---|---|---|
| Erythromycine | 3 | 128 | 0.5 | 8 | 2 | 16 | 4 | 256 | <0.0005 |
| 4 | 256 | 4 | 16 | 32 | 64 | 32 | 512 | 0.031 | |
| 5 | 512 | 64 | 32 | 64 | 256 | 128 | >1024 | 4 | |
| 6 | 512 | 128 | 64 | 256 | 512 | 128 | >1024 | 2 | |
| Josamycine | 3 | 64 | 4 | 16 | 4 | 64 | 16 | 64 | 0.0019 |
| 4 | 512 | 16 | 32 | 64 | 128 | 64 | 128 | 1 | |
| 5 | 1024 | 128 | 64 | 256 | 256 | 128 | 512 | 8 | |
| 6 | >1024 | 512 | 128 | 512 | 1024 | 256 | 1024 | 16 | |
| Azithromycine | 3 | 8 | 0.031 | 0.25 | 0.031 | 0.5 | 0.5 | 8 | <0.0005 |
| 4 | 32 | 1 | 0.5 | 1 | 4 | 1 | 16 | 0.25 | |
| 5 | 64 | 4 | 4 | 4 | 16 | 4 | 32 | 1 | |
| 6 | 64 | 4 | 4 | 8 | 32 | 4 | 64 | 2 | |
| Roxithromycine | 3 | 512 | 16 | 16 | 4 | 64 | 16 | 512 | <0.0005 |
| 4 | 1024 | 16 | 64 | 32 | 256 | 64 | 1024 | 4 | |
| 5 | 1024 | 128 | 128 | 256 | 512 | 256 | >1024 | 16 | |
| 6 | 1024 | 512 | 256 | 256 | 1024 | 256 | >1024 | 16 | |
| Spiramycine | 3 | >1024 | 16 | 64 | 4 | 256 | 32 | 512 | <0.0005 |
| 4 | >1024 | 64 | 128 | 256 | 512 | 128 | 256 | 1 | |
| 5 | 1024 | 256 | 128 | 512 | 1024 | 512 | 512 | 1 | |
| 6 | >1024 | 256 | 256 | 1024 | 1024 | 1024 | 512 | 1 | |
| clarithromycine | 3 | 32 | 4 | 4 | 2 | 8 | 1 | 512 | 0.0039 |
| 4 | 128 | 8 | 8 | 4 | 64 | 8 | 1024 | 16 | |
| 5 | 128 | 16 | 32 | 32 | 256 | 64 | 1024 | 16 | |
| 6 | 128 | 32 | 32 | 64 | 512 | 64 | 1024 | 32 | |
| Dirithromycine | 3 | 64 | 8 | 8 | 4 | 16 | 4 | 128 | <0.0005 |
| 4 | 512 | 16 | 16 | 8 | 64 | 8 | 256 | 2 | |
| 5 | 512 | 16 | 16 | 16 | 64 | 32 | 512 | 4 | |
| 6 | 512 | 32 | 16 | 16 | 128 | 32 | 512 | 8 | |
| Tylosine | 3 | 512 | 16 | 128 | 4 | 128 | 32 | 128 | 4 |
| 4 | >1024 | 256 | 256 | 128 | 1024 | 256 | 256 | 32 | |
| 5 | >1024 | 512 | 512 | 512 | >1024 | 512 | 512 | 64 | |
| 6 | >1024 | 512 | 512 | 512 | >1024 | 512 | 1024 | 128 |
a E. coli ATCC 25922. b K. pneumoniae ATCC 13883. c C. koseri IP8294. d E. cloacae DSM 129. e K. aerogenes ATCC 13048. f K. aerogenes 289 (MDR). g E. coli AG100A_pUC18.
Considering macrolides as OM-impermeable antibiotics, their antibacterial spectrum is restricted mostly to Gram-positive organisms [39]. Potentiation of these antibiotics by compound 3 was prevalent against all wild-type (except AG100A puc) Gram-negative bacteria tested. The mechanism of action of macrolides is well known to involve inhibition of bacterial protein synthesis, and they were rendered active against Gram-negative bacteria by the adjuvant allowing them to overcome the OM barrier. Since compound 3 (with a farnesyl and a long polyamine chain (spermine)) is more potent than compound 4 (with a ramified polyamine group) and derivatives 5 and 6 (with a shorter geranyl moiety), it clearly appears that both the presence of a long hydrophobic carbon chain and a highly charged linear polyamine group bestow preferential membrane interactions relative on these compounds. All these assumptions tend to suggest that these compounds can disrupt the OM integrity facilitating the entrance of hydrophobic antibiotics. It is also noteworthy that the ability of compound 3 to enhance macrolide’s activity was further evaluated against MDR clinical isolate Ea289 with similar success than toward sensitive ones.
The value of the Fractional Inhibitory Concentration (FIC index) as a predictor of synergy was investigated using the various macrolides as antibacterial agents combined with compound 3 in fully blind experiments against numerous different bacterial strains (Table 5). Under the conditions used, except for PA01 strain for which no synergy was observed whatever the considered macrolide, synergies were encountered with FIC index <0.25 in numerous cases, demonstrating the strong effect of compound 3 to restore the antibacterial activity of macrolides against Gram-negative bacteria.
Table 5.
Fractional Inhibitory Concentration Index (FICI) obtained for interactions of compound 3 with eight different macrolides against various Gram-negative bacteria and classified as follows: FICI ≤ 0.5 = synergistic (yellow); 0.6 < FICI ≤ 0.9 = additive (green); and 1 < FICI ≤ 3.9 = indifferent (red). The variation of the color from yellow to green is related to the importance of the synergy (from higher to lower synergy).
| Strain | FICI | |||||||
|---|---|---|---|---|---|---|---|---|
| Erythro | Josa | Azithro | Roxithro | Spira | Clarithro | Dirithro | Tylo | |
| Pa01 | 0.65 | 0.46 | 0.46 | 0.90 | 1.40 | 0.46 | 0.46 | 0.90 |
| E.c. a | 0.20 | 0.20 | 0.20 | 0.20 | 0.23 | 0.23 | 0.33 | 0.23 |
| K.p. b | 0.26 | 0.22 | 0.22 | 0.23 | 0.33 | 0.23 | 0.33 | 0.33 |
| C.k. c | 0.21 | 0.20 | 0.20 | 0.22 | 0.20 | 0.22 | 0.33 | 0.20 |
| E.cl. d | 0.22 | 0.26 | 0.21 | 0.26 | 0.45 | 0.21 | 0.33 | 0.33 |
| K.a. e | 0.11 | 0.13 | 0.11 | 0.13 | 0.13 | 0.10 | 0.16 | 0.13 |
| Ea289 f | 0.35 | 0.16 | 0.23 | 0.60 | 0.60 | 0.60 | 0.23 | 0.23 |
a E. coli ATCC 25922. b K. pneumoniae ATCC 13883. c C. koseri IP8294. d E. cloacae DSM 129. e K. aerogenes ATCC 13048. f K. aerogenes 289 (MDR).
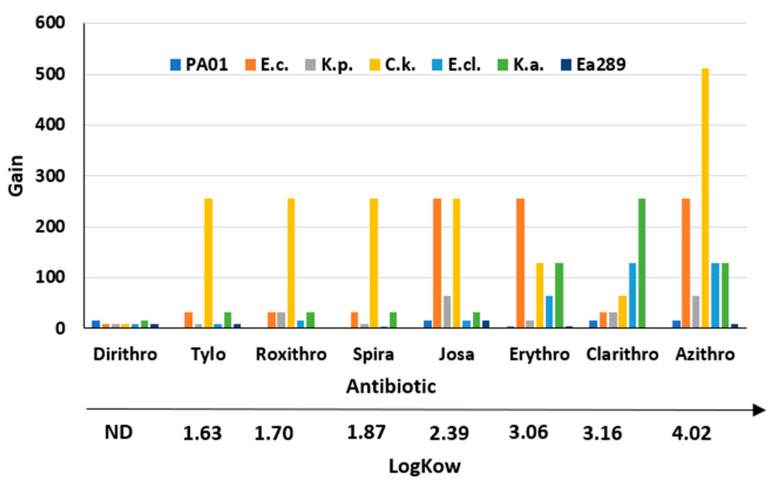
It is interesting to note that some macrolides, such as josamycin, erythromycin, clarithromycin, and azithromycin, are better than others—e.g., dirithromycine, tylosine, roxithromycine, and spiramycine—when combined with compound 3 against the considered bacterial strains. Thus, by considering the gain obtained (as the ratio of the MIC of the macrolide alone to the MIC of the macrolide combined with 3 used at a 10 µM concentration), we clearly notice a strong correlation with the LogKow (values found in PubChem for all the tested macrolides) of the macrolides, commonly used as a measure of hydrophobicity, since, overall, the higher the lipophilicity of the compound, the higher the obtained gain (Figure 1 and Figure 2).
Figure 2.
Correlation of the gain factor observed toward numerous Gram-negative bacteria depending on the LogKow value of the considered macrolide antibiotic.
2.5. Mechanism of Action of Compound 3
Inner Membrane Depolarization Assay
The most efficient adjuvant (Cpd 3) was evaluated for its potent ability to disrupt the proton gradient of the bacterial inner membranes of the Gram-negative bacteria [40]. Thus, to monitor the phenomenon, DiSC3(5) assay was used to measure the electrical potential gradient across the inner membrane. DiSC3(5) is a cationic membrane permeable fluorescent dye which can build up on hyperpolarized membrane and translocate into the lipid bilayer. If compound 3 interferes with the inner membrane, then it will lead to a membrane depolarization with the dye being consequently released into the external environment. The increase of fluorescence is then recorded and subsequently quantified.
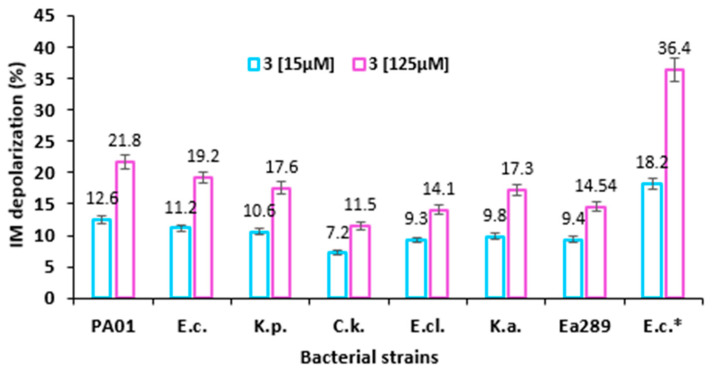
Figure 3 shows the dose-dependent increase (at two different concentrations, 15 and 125 µM) in the percentages of depolarization (calculated on the basis of untreated controls) of the inner membrane resulting from its alteration by compound 3. Interestingly, no significant differences were observed between all the considered Gram-negative strains whatever the concentration tested, except for AG100A_pUC18 which does not present any AcrAB efflux pumps expression. In the latter case, the obtained values were almost twofold higher compared with the rest of the strains tested at the same concentration of compound 3. The other tested concentrations of compound 3 on the inner membrane depolarization of various Gram-negative bacteria are presented in Figure S2.
Figure 3.
Inner membrane depolarization of various Gram-negative bacteria (PA01, E. coli ATCC 25922, K. pneumoniae ATCC 13883, C. koseri IP8294, E. cloacae DSM 129, K. aerogenes ATCC 13048, K. aerogenes 289 (MDR), and E. coli AG100A_pUC18) by evaluating DiSC3(5) fluorescence recorded after 5 minutes in the presence of compound 3 at 15 µM and 125 µM. * Refers to E. coli AG100A_pUC18. The results represent the average plus SD of three independent experiments.
2.6. ATP Efflux Measurement
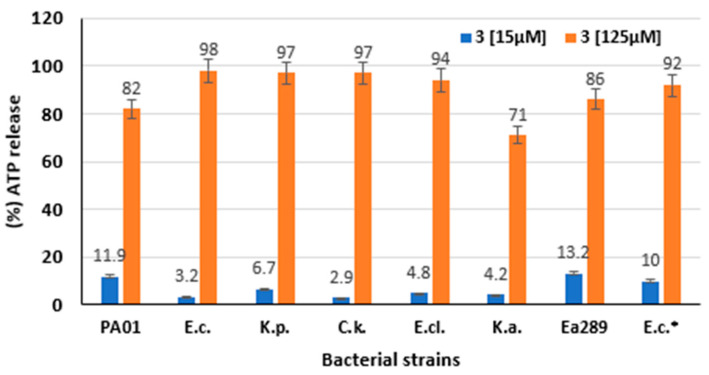
A bioluminescence method was utilized to determine the behavior of compound 3 on the intracellular pool of bacterial ATP. Thus, the external concentration of ATP was used as a reporter reflecting the permeabilizing effect of 3 (Figure 4) along with providing a dose–response curve (Figure S3) [41,42]. Thus, 3 used at a 125 µM concentration dose dramatically disrupted the Gram-negative bacterial membranes after 1 min as observed by intracellular ATP release kinetics, which was similar to that observed for the positive control polymyxin B (Figure 4). Conversely, no significant effect was found by using spermine as a polyamine negative control during the test time). Lower but significant ATP efflux was observed after 1 min for compound 3 used at a 15 µM concentration with 2.9 to 13.2% ATP efflux release relative to the CTAB positive control depending on the nature of the considered bacteria, respectively. As illustrated in Figure 4, compound 3 led to a significant level of ATP release against the multidrug-resistant Ea289 strain even at low concentrations, since the percentage of ATP detected was up to twofold higher compared with that of the other strains (see Figure S3 for dose-dependent data).
Figure 4.
ATP release levels measurement in the presence of compound 3. ATP release levels of various Gram-negative bacteria (PA01, E. coli ATCC 25922, K. pneumoniae ATCC 13883, C. koseri IP8294, E. cloacae DSM 129, K. aerogenes ATCC 13048, K. aerogenes 289 (MDR), and E. coli AG100A_pUC18) evaluated after 1 minute by bioluminescence in the presence of compound 3 at 15 µM and 125 µM. In each case, polymyxin B (250 µM) was used as a positive control to quantify the maximum level of ATP efflux. * Refers to E. coli AG100A_pUC18.
2.7. Outer Membrane Permeabilization
The nitrocefin hydrolysis method was used to evaluate the effect of 3 on the integrity of the outer membranes of the different Gram-negative bacteria. This assay relies upon the hydrolysis of the chromogenic β-lactam nitrocefin periplasmic beta-lactamases, leading to a change in color from yellow to red, relating color change to the degree of integrity of the outer membrane.
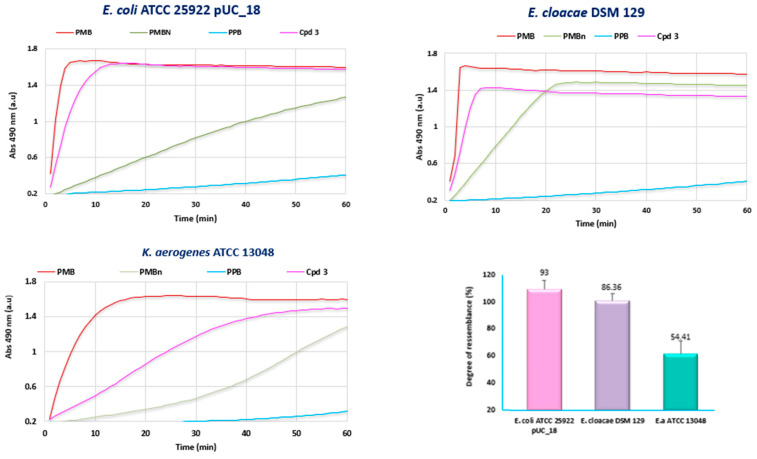
In this context, Figure 5 presents the results of our investigations against some selected Gram-negative bacteria (producing β-lactamases) (E. coli ATCC25922 pUC_18, E. cloacae DSM 129, and K. aerogenes ATCC 13048) in the presence and absence of compound 3 or two positive controls (Polymyxin B (PmB) and Polymyxin Nona PmNona).
Figure 5.
Outer membrane permeabilization of chosen Gram-negative bacterial strains by evaluating the rate of nitrocefin hydrolysis in the presence of PPB (Potassium Phosphate Buffer), PMB, PMBn, and compound 3 at 125 µM.
At a high concentration of 125 µM, compound 3 showed a strong effect on the OM permeability compared with polymyxin B. Interestingly, the increase of the nitrocefin hydrolysis rate in the presence of 3 was highly dependent on the considered Gram-negative bacteria. This rate ranged from 54% to 93% compared with polymyxin B suggesting that the behavior of 3 with respect to the binding of the outer membrane was not similar. The results of the nitrocefin test performed against all the tested Gram-negative strains in this study are presented in Supplementary Data (Figure S4).
3. Discussion
Infectious diseases caused by Gram-negative bacterial resistant strains represent a major health concern. Studies have demonstrated that in most multidrug-resistant strains, the alteration of the cell envelope permeability is frequently reported. In previous works, we demonstrated that the OM could play a major role in the susceptibility of these bacteria to antibiotics. Gram-negative bacteria can proceed to the modification of lipid A or proteins in OM composition leading to the resistance or antibiotic susceptibility of the considered species [13]. In a previous study, we determined that the combination of a farnesyl spermine compound 3 used at concentrations ranging from 2.5 to 10 µM, in the presence of doxycycline or minocycline, leads to a significant decrease of P. aeruginosa antibiotic resistance towards these antibiotics [37]. In the context of our continuing studies, we have demonstrated a stronger synergetic effect using a combination of compound 3 and a macrolide leading to an increase in Gram-negative bacteria susceptibility to this antibiotic family. Interestingly, the in vitro experiments presented different antimicrobial activity profiles depending on the considered bacterial strain, which could suggest that different hydrophobicity levels of the tested macrolides lead to different interaction pathways with the outer membrane of these pathogens.
For further understanding of compound 3 behavior, its mechanism of action was more precisely investigated, demonstrating the strongest antimicrobial activity among all the parent derivatives tested. On the basis of the outer membrane permeabilization assay, our data suggested that compound 3 disrupts the outer membrane integrity of Gram-negative bacteria more strongly compared with polymyxin nona (used as an internal control). More interestingly, the behavior of compound 3 toward the outer membrane of the different tested strains was highly variable. This suggests that the differences encountered may be due to variation in Gram-negative bacteria outer membrane composition, implying that the role of lipid A could strongly modify the bacterial adaptation to withstand external stresses [13]. This study also demonstrated that compound 3 has a weak effect on the depolarization of the bacterial inner membrane and that this phenomenon occurs in a dose-dependent manner. Interestingly, this depolarization was twofold higher in the case of AG100A_pUC18, which does not exhibit efflux pumps. The same result was obtained by measuring ATP efflux in the presence of compound 3 where no significant differences can be noticed between the different strains, except for Ag100A_pUC18.
Taken together, these results suggest that compound 3 presents a weak effect on the inner membrane depolarization as evidenced by a weak ATP efflux level, but it strongly disrupts the integrity of the outer membrane of Gram-negative pathogens.
4. Methods and Materials
All the solvents were purified according to reported procedures, and the reagents used were commercially available. Methanol, ethyl acetate, and dichloromethane were purchased from Sigma-Aldrich (St. Quentin Fallavier, France) and used without further purification. Column chromatography was performed on Merck silica gel (70–230 mesh). 1H NMR and 13C NMR spectra were recorded in MeOD on a Bruker AC 300 spectrometer working at 300 MHz and 75 MHz, respectively (The usual abbreviations were used: s: singlet, d: doublet, t: triplet, q: quadruplet, and m: multiplet). All chemical shifts are given in ppm. Mass spectroscopy analysis was performed by the Spectropole Laboratory (Marseille, France). The purity of the compounds was checked by analytical HPLC (C18 column, eluent CH3CN-water-TFA (90:10:0.025, v/v/v), 0.5–1 mL/Min) with PDA detector spanning from 210 nm to 310 nm. All compounds possessed purity above 95%, as determined by analytical HPLC-PDA at 210 nm.
4.1. Procedure for the Synthesis of Polyaminoisoprenyl Derivatives 3–6
Synthesis of Compound 3
To a solution of spermine (450 mg, 2.27 mmol) and triethylamine (450 µL, 4.5 mmol) in distilled tetrahydrofuran (THF) (10 mL) was added dropwise farnesyl chloride 1 (480 mg, 2 mmol) in THF (15 mL). The reaction mixture was stirred at room temperature for 24 h and evaporated to dryness. The crude residue was purified by column chromatography (eluant CH2Cl2/MeOH/conc. NH4OH, 7:3:1) to afford the pure desired compound in 64% yield as a mixture of isomers. Yellow solid; 1H NMR (MeOD, 300 MHz): δ = 5.28–4.93 (m, 3H), 2.93–2.57 (m, 14H), 2.19–1.92 (m, 10H), 1.87–1.63 (m, 23H). 13C (MeOD, 75 MHz): δ = 140.06, 139.44, 136.28, 136.25, 136.16, 136.13, 132.37, 132.13, 132.10, 125.94, 125.42, 125.38, 125.21, 123.15, 121.80, 54.58, 52.48, 52.25, 50.66, 50.52, 48.77, 48.24, 48.01, 47.63, 41.28, 41.18, 41.12, 40.94, 40.90, 40.80, 40.62, 33.26, 32.99, 30.35, 30.11, 28.53, 28.21, 27.84, 27.66, 27.43, 26.09, 26.04, 25.41, 23.84, 23.79, 17.90, 17.86, 16.61, 16.48, 16.25, and 16.21. C25H50N4 MS (ESI+) m/z 407.41 (100%, [M + H]+), and cald. 407.703.
All the other compounds 4–6 were synthesized according to a previous reported procedure [36] (see Figure S1).
4.2. Bacterial Strains
For this study, we used some of the most common Gram-negative bacteria involved in severe infections, primarily reference strains (PA01, E. coli ATCC 25922, K. pneumoniae ATCC 13883, C. Koseri IP8294, E. cloacae DSM 129, and K. aerogenes ATCC 13048) and a multiresistant strain K. aerogenes 289, as well as an E. coli β-lactamase-producing strain AG100A_pUC18. These strains were stored in 15% (v/v) glycerol at −80 °C for cryo-protection and sub-cultured overnight in Mueller–Hinton broth 2 (MH II) at 37 °C for inoculum preparation.
4.3. Transfer of Plasmid pUC-18 into Escherichia coli Strains
The different strains of E. coli were made competent by a calcium chloride treatment then transformed with the pUC18 plasmid by a heat shock. A single colony of AG100A was inoculated into 10 mL of Luria–Bertani (LB) medium and bacteria were grown until OD600nm = 0.4 at 37 °C with shaking (180 rpm). The cultures were transferred into a centrifuge bottle, placed on ice for 20 min and centrifuged at 4000× g for 10 min at 4 °C. The pellets were resuspended in 5 mL of CaCl2 at 50 mM and incubated in ice for 1 h. Bacteria were centrifuged again, and the pellets were gently resuspended in 200 µL of CaCl2 50 mM and glycerol 15%. Then, 100 ng of pUC18 was added at 100 µL of the competent strain and the cells were incubated in ice for 20 min. They were then incubated 2 min at 42 °C and transferred in ice. Subsequently, 900 µL of LB was added, and the tubes were incubated at 37 °C with shaking (180 rpm) for 1 hour. Dilutions were then prepared and spread on LB agar plates containing 100 µg/mL of ampicillin. Colonies were selected after overnight incubation at 37 °C and PCR tests were performed for verification by amplifying the gene coding for the β-lactamase on pUC18.
4.4. Antibiotics
A large panel of antibiotics belonging to the macrolide family—such as erythromycin, josamycin, roxithromycin, azithromycin, spiramycin, clarithromycin, dirithromycin, and tylosin—was used in this study. All antibiotics were purchased from Sigma (St. Quentin Fallavier, France) and dissolved in water, dimethyl sulfoxide (DMSO), or ethanol as indicated.
4.5. MIC Determination of Macrolides and Polyaminoisoprenyl Derivatives
The minimal inhibitory concentration (MIC) is defined as the lowest concentration of an antibiotic able to neutralize the majority (99.9%) of a bacterial inoculum. Susceptibilities to macrolides and compounds 3–6 were determined in sterile 96-well microplates by using the standard broth dilution method in accordance with the recommendations of the Comité de l’Antibiogramme de la Société Française de Microbiologie (CA-SFM) [43]. The stock solutions of macrolide antibiotics were freshly prepared at a 51.2 mg/mL concentration for each experiment in water, ethanol, or dimethyl sulfoxide (DMSO) as indicated. Briefly, the MICs were determined with an inoculum of 105 CFU in 200 µL of MH2 broth containing twofold serial dilutions ranging from 1024 µg/mL to 2 µg/mL of each molecule. The MIC was defined as the lowest concentration of drug that completely inhibited visible growth after incubation for 18 h at 37 °C. All MIC determinations were repeated in triplicate in independent experiments.
4.6. Determination of MICs of Macrolides in the Presence of Synergizing Compounds 3–6
The antimicrobial activities of macrolides in combination with compounds 3–6 at a 10 µM concentration were evaluated in sterile 96-well microplates. A twofold serial dilution of the drugs (from 1024 μg/mL to 2 μg/mL) was performed from the starting solution. In each column of the microplate, the concentration of the adjuvants was set at 10 μM/well. The bacterial suspension was prepared from colonies grown overnight. The concentration was adjusted to 105 CFU/well. The MIC of each combination was determined after 18 h of incubation at 37 °C. All MIC determinations were repeated at least three times in independent experiments. The gain was defined as the ratio of the MIC of each macrolide antibiotic to its MIC determined in the presence of each adjuvant.
4.7. Chequerboard Assay/Fractional Inhibitory Concentration Index (FICI)
To determine the interaction and to evaluate the combined effect of the considered drug in the presence of compound 3, a chequerboard method was used. This test allows for the determination of the MIC and FICI values at the same time. This test was performed in 96-well microplates. The amount of 50 µL of a twofold serial dilution of each macrolide was added in the lines. Then, 50 µL of the different concentrations of compound 3 ranging from 50 µM to 0.78 µM was dispensed in the columns. Then, 100 µL of the bacterial suspension containing 105 CFU/mL was added to the different wells and the plates were incubated at 37 °C for 24 h.
The combination effects were evaluated by the sum of FICIs of the macrolide–compound 3 combination.
Four types of effects were classified as follows: FICI ≤ 0.5 = synergistic; 0.6 < FICI ≤ 0.9 = additive; 1 < FICI ≤ 3.9 = indifferent; and FICI > 4.0 = antagonistic
4.8. Outer Membrane Permeabilization Assay
Nitrocefin was used as a chromogenic substrate of periplasmic β-lactamase to measure the outer membrane permeabilization. The nitrocefin hydrolysis assay is a colometric assay wherein a color change from yellow to red occurs when the chromogenic β-lactam is efficiently hydrolyzed by periplasmic β-lactamases. This test was determined on the different Gram-negative bacteria (PA01, E. coli ATCC 25922, K. pneumoniae ATCC 13883, C. Koseri IP8294, E. cloacae DSM 129, K. aerogenes ATCC 13048, K. aerogenes 289, and AG100A_pUC18) to investigate the effect of the polyaminoisoprenyl derivatives on the outer membrane.
After an overnight culture of the different bacteria at 37 °C, 100 µL of each suspension was added to 10 mL of MHII broth, except for AG100A_pUC18 whose suspension was supplemented with 100 µg/mL of ampicillin to maintain the pUC 18 plasmid. It is noteworthy that for PA01 and E. aerogenes ATCC 13048 suspensions, 0.001 µg/mL of imipenem was added when the cultures reached the mid-logarithmic phase (OD600 = 0.5) to induce the β-lactamase production. The cells were then recovered by centrifugation (3600× g for 20 min at 20 °C) and washed twice with 20 mM potassium phosphate buffer (pH 7.2) and 1 mM MgCl2 (PPB). After a second centrifugation, the pellet was resuspended and adjusted to an OD600 of 0.375. Then, 100 µL of each bacterial suspension was mixed with 50 µL of a solution of compounds 3–6 at a concentration of 128 µM already set up in a 96-well microplate. Polymyxin B (PMB) and polymyxin Nona (PMBn) were used as positive controls, and PPB was used as a negative control. Finally, 50 µL of nitrocefin was added to obtain a final concentration of 50 µg/mL. Nitrocefin hydrolysis was monitored by measuring the increase in absorbance at 490 nm using a M200 Pro Tecan spectrophotometer for 1 h with a 1-min interval between each measurement. Experiments were performed in triplicate.
4.9. Membrane Depolarization Assay
The different Gram-negative strains were grown in MH II broth for 24 h at 37 °C. After reaching an OD600 nm of 0.5, cells were centrifuged (3600× g for 20 min at 20 °C) and washed twice in Hepes (5 mM) (pH = 7.2) supplemented with sucrose (250 mM final concentration) and MgCl2 (25 mM final concentration). The fluorescent dye 3,3′-diethylthiacarbocyanine iodide DiSC3(5) was added to a final concentration of 5 µM and was incubated with the suspensions for 5 min at 37 °C to allow the dye incorporation into the polarized membranes. Then, 10 µL of compound 3 (the most efficient compound) was added to 90 µL of the fluorescent suspensions at different concentrations ranging from 250 µM to 7.8 µM. Fluorescence measurements were recorded after 1 min, 5 min, 10 min, and 15 min (excitation wavelength 622 nm, emission wavelength 690 nm).
The difference in the relative fluorescence values (RFU) from the control containing only buffer and the control containing bacteria treated only with cetyltrimethylammonium bromide (CTAB 1%) was chosen as the maximum level of depolarization. Assays were performed in three independent experiments.
4.10. ATP Efflux Measurement
Different solutions of compound 3 were prepared in twice-distilled water with a concentration ranging from 250 to 3.91 µM. The different Gram-negative suspensions were prepared in MH II broth and were incubated at 37 °C. Then, 90 µL of each bacterial suspension was added to 10 µL of a compound 3 solution and shaken for 20 s in the incubator at 37 °C. Subsequently, 50 µL of Luceferin–Luceferase reagent (Yelen, France) was added to the mixture, and luminescent signal quantified with an Infinite M200 microplate reader (Tecan, Männedorf, Switzerland) for five seconds. ATP concentration was quantified by internal sample addition. Polymyxin B (250 µM) was used as a positive control to quantify the maximum level of ATP efflux. This assay was performed in three independent experiments.
4.11. Cytoxicity Assays
Cytotoxicity assessment was performed on the referenced Chinese Hamster Ovary cell line (CHO-K1, ATCC-LGC Promochem, Molsheim, France). Cells were maintained in McCoy’s 5A medium (Sigma) supplemented with 10% foetal calf serum, 1 mM glutamine, and penicillin–streptomycin (100 U·mL−1 and 10 μg·mL−1, respectively), and incubated at 37 °C in a humidified atmosphere containing 5% CO2. The cytotoxic effects of compounds were assessed by the colorimetric WST-1 cell proliferation assay. Briefly, a range of compounds concentrations from 30 µM to 1200 µM was incorporated in triplicate cultures, and cells were incubated at 37 °C for 24 h. At the end of the incubation period, cultures were submitted to three successive washes in phosphate buffer saline (PBS) and incubated in fresh culture medium containing 10% WST-1 for an additional 30 min. Cell viability was evaluated by the assessment of WST-1 absorbance at 450 nm in a microplate spectrophotometer MRX1 II (Dynex Technologies, Chantilly, VA, USA). The Inhibitory Concentration 50% (IC50) was chosen to evaluate the cytotoxicity of compounds. IC50 was defined as the concentration of compounds that induced a 50% decrease of viable cells.
5. Conclusions
An original strategy has been developed affording new polyaminoisoprenyl compounds which exhibited a strong effect on the level of macrolide antibiotics susceptibility for resistant Gram-negative bacterial strains. This activity was correlated to the level of hydrophobicity of the antibiotics as well as to the ability of the polyaminoisoprenyl derivatives to alter bacterial outer membrane integrity. For the first time, we demonstrated that weak membrane perturbation is ideal for designing an intrinsically inactive, non-toxic adjuvant for non-specific potentiation of antibiotics. Whilst further testing is required before these compounds can be approved as antibiotic adjuvants for therapeutic use, outer membrane proteins are proving to be promising targets offering hope in the ongoing battle against antimicrobial resistance. Studies are now underway to determine if this restoration of antibiotic susceptibility occurs also by a direct interaction of the molecule with the efflux pump or by another mechanism.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms232012457/s1.
Author Contributions
Formal analysis, J.M.B. (Jean Michel Brunel); Funding acquisition, J.M.B. (Jean Michel Brunel); Investigation, A.T. and J.M.B. (Jean Michel Brunel); Methodology, A.T., J.M.B. (Jean Michel Bolla) and N.K.; Project administration, J.M.B. (Jean Michel Brunel); Supervision, J.M.B. (Jean Michel Brunel); Writing—original draft, J.M.B. (Jean Michel Brunel). All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Aslam B., Wang W., Arshad M.I., Khurshid M., Muzammil S., Rasool M.H., Nisar M.A., Alvi R.F., Aslam M.A., Qamar M.U., et al. Antibiotic resistance: A rundown of a global crisis. Infect. Drug Resist. 2018;11:1645–1658. doi: 10.2147/IDR.S173867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Livermore D.M. The need for new antibiotics. Clin. Microbiol. Infect. 2004;10((Suppl. 4)):1–9. doi: 10.1111/j.1465-0691.2004.1004.x. [DOI] [PubMed] [Google Scholar]
- 3.Payne D.J. Microbiology. Desperately seeking new antibiotics. Science. 2008;321:1644–1645. doi: 10.1126/science.1164586. [DOI] [PubMed] [Google Scholar]
- 4.Aminov R.I. The role of antibiotics and antibiotic resistance in nature. Environ. Microbiol. 2009;11:2970–2988. doi: 10.1111/j.1462-2920.2009.01972.x. [DOI] [PubMed] [Google Scholar]
- 5.Saleem M., Nazir M., Ali M.S., Hussain H., Lee Y.S., Riaz N., Jabbar A. Antimicrobial natural products: An update on future antibiotic drug candidates. Nat. Prod. Rep. 2010;27:238–254. doi: 10.1039/B916096E. [DOI] [PubMed] [Google Scholar]
- 6.Peterson E., Kaur P. Antibiotic resistance mechanisms in bacteria: Relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogen. Front. Microbiol. 2018;9:2928. doi: 10.3389/fmicb.2018.02928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vivas R., Barbosa A.A.T., Dolabela S.S., Jain S. Multidrugresistant bacteria and alternative methods to control them: An overview. Microb. Drug Resist. 2019;25:890–908. doi: 10.1089/mdr.2018.0319. [DOI] [PubMed] [Google Scholar]
- 8.De Angelis G., D’Inzeo T., Fiori B., Spanu T., Sganga G. Burden of antibiotic resistant gram negative bacterial infections: Evidence and limits. J. Med. Microb. Diagn. 2014;3:1000132. [Google Scholar]
- 9.Oduro-Mensah D., Obeng-Nkrumah N., Bonney E.Y., Oduro-Mensah E., Twum-Danso K., Osei Y.D., Sackey S.T. Genetic characterization of TEM-type ESBL-associated antibacterial resistance in Enterobacteriaceae in a tertiary hospital in Ghana. Ann. Clin. Microbiol. Antimicrob. 2016;15:29. doi: 10.1186/s12941-016-0144-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagenlehner F.M.E., Dittmar F. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Eur. Urol. 2022;399:629–655. doi: 10.1016/j.eururo.2022.08.023. [DOI] [PubMed] [Google Scholar]
- 11.Cepas V., López Y., Muñoz E., Rolo D., Ardanuy C., Martí S., Xercavins M., Horcajada J.P., Bosch J., Soto S.M. Relationship between biofilm formation and antimicrobial resistance in gram-negative bacteria. Microb. Drug Resist. 2019;25:72–79. doi: 10.1089/mdr.2018.0027. [DOI] [PubMed] [Google Scholar]
- 12.Douafer H., Andrieu V., Phanstiel O., Brunel J.M. Antibiotic Adjuvants: Make Antibiotics Great Again! J. Med. Chem. 2019;62:8665–8681. doi: 10.1021/acs.jmedchem.8b01781. [DOI] [PubMed] [Google Scholar]
- 13.Troudi A., Pages J.M., Brunel J.M. Chemical Highlights Supporting the Role of Lipid A in Efficient Biological Adaptation of Gram-Negative Bacteria to External Stresses. J. Med. Chem. 2021;64:1816–1834. doi: 10.1021/acs.jmedchem.0c02185. [DOI] [PubMed] [Google Scholar]
- 14.Clifton L.A., Skoda M.W., Le Brun A.P., Ciesielski F., Kuzmenko I., Holt S.A., Lakey J.H. Effect of divalent cation removal on the structure of Gram-negative bacterial outer membrane models. Langmuir. 2015;31:404–412. doi: 10.1021/la504407v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Decad G.M., Nikaido H. Outer membrane of Gram-negative bacteria. XII. Molecular-sieving function of cell wall. J. Bacteriol. 1976;128:325–336. doi: 10.1128/jb.128.1.325-336.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nikaido H. Outer membrane of Salmonella typhimurium transmembrane diffusion of some hydrophobic substances. Biochim. Biophys. Acta Biomembr. 1976;433:118–132. doi: 10.1016/0005-2736(76)90182-6. [DOI] [PubMed] [Google Scholar]
- 17.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Shea R., Moser H.E. Physicochemical properties of antibacterial compounds: Implications for drug discovery. J. Med. Chem. 2008;51:2871–2878. doi: 10.1021/jm700967e. [DOI] [PubMed] [Google Scholar]
- 19.Tommasi R., Brown D.G., Walkup G.K., Manchester J.I., Miller A.A. ESKAPEing the labyrinth of antibacterial discovery. Nat. Rev. Drug Discov. 2015;14:529–542. doi: 10.1038/nrd4572. [DOI] [PubMed] [Google Scholar]
- 20.Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klobucar K., French S., Côté J.P., Howes J.R., Brown E.D. Genetic and chemical-genetic interactions map biogenesis and permeability determinants of the outer membrane of Escherichia coli. mBio. 2020;11:e00161-20. doi: 10.1128/mBio.00161-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barker C.A., Farha M.A., Brown E.D. Chemical genomic approaches to study model microbes. Chem. Biol. 2010;17:624–632. doi: 10.1016/j.chembiol.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 23.Specht K.M., Shokat K.M. The emerging power of chemical genetics. Curr. Opin. Cell Biol. 2002;14:155–159. doi: 10.1016/S0955-0674(02)00317-4. [DOI] [PubMed] [Google Scholar]
- 24.Repaske R. Lysis of Gram-negative organisms and the role of versene. Biochim. Biophys. Acta. 1958;30:225–232. doi: 10.1016/0006-3002(58)90044-1. [DOI] [PubMed] [Google Scholar]
- 25.Vaara M. Agents that increase the permeability of the outer membrane. Microbiol. Rev. 1992;56:395–411. doi: 10.1128/mr.56.3.395-411.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corbett D., Wise A., Langley T., Skinner K., Trimby E., Birchall S., Dorali A., Sandiford S., Williams J., Warn P., et al. Potentiation of antibiotic activity by a novel cationic peptide: Potency and spectrum of activity of SPR741. Antimicrob. Agents Chemother. 2017;61:e00200-17. doi: 10.1128/AAC.00200-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.French S., Farha M.A., Ellis M.J., Sameer Z., Côté J.-P., Cotroneo N., Lister T., Rubio A., Brown E.D. Polymyxin B analog SPR741 potentiates antibiotics against Gram-negative bacteria and uniquely perturbs the outer membrane. ACS Infect. Dis. 2020;6:1405–1412. doi: 10.1021/acsinfecdis.9b00159. [DOI] [PubMed] [Google Scholar]
- 28.Hancock R.E.W. Peptide antibiotics. Lancet. 1997;349:418–422. doi: 10.1016/S0140-6736(97)80051-7. [DOI] [PubMed] [Google Scholar]
- 29.Schindler M., Osborn M.J. Interaction of divalent cations and polymyxin B with lipopolysaccharide. Biochemistry. 1979;18:4425–4430. doi: 10.1021/bi00587a024. [DOI] [PubMed] [Google Scholar]
- 30.Vaara M., Viljanen P. Binding of polymyxin B nonapeptide to Gram-negative bacteria. Antimicrob. Agents Chemother. 1985;27:548–554. doi: 10.1128/AAC.27.4.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dias C., Pais J.P., Nunes R., Blázquez-Sánchez M.T., Marquês J.T., Almeida A.F., Serra P., Xavier N.M., Vila-Viçosa D., Machuqueiro M., et al. Sugar-based bactericides targeting phosphatidylethanolamine-enriched membranes. Nat. Commun. 2018;19:4857. doi: 10.1038/s41467-018-06488-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vázquez-Laslop N., Mankin A.S. How Macrolide Antibiotics Work. Trends Biochem. Sci. 2018;43:668–684. doi: 10.1016/j.tibs.2018.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gomes C., Martínez-Puchol S., Palma N., Horna G., Ruiz-Roldán L., Pons M.J., Ruiz J. Macrolide resistance mechanisms in Enterobacteriaceae: Focus on azithromycin. Crit. Rev. Microbiol. 2017;43:1–30. doi: 10.3109/1040841X.2015.1136261. [DOI] [PubMed] [Google Scholar]
- 34.Arsic B., Barber J., Čikoš A., Mladenovic M., Stankovic N., Novak P. 16-membered macrolide antibiotics: A review. Int. J. Antimicrob. Agents. 2018;51:283–298. doi: 10.1016/j.ijantimicag.2017.05.020. [DOI] [PubMed] [Google Scholar]
- 35.Borselli D., Lieutaud A., Thefenne H., Garnotel E., Pagès J.-M., Brunel J.M., Bolla J.-M. Polyamino-isoprenic derivatives block intrinsic resistance of P. aeruginosa to doxycycline and chloramphenicol in vitro. PLoS ONE. 2016;11:e0154490. doi: 10.1371/journal.pone.0154490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lieutaud A., Pieri C., Bolla J.M., Brunel J.M. New Polyaminoisoprenyl Antibiotics Enhancers against Two Multidrug-Resistant Gram-Negative Bacteria from Enterobacter and Salmonella Species. J. Med. Chem. 2020;63:10496–10508. doi: 10.1021/acs.jmedchem.0c01335. [DOI] [PubMed] [Google Scholar]
- 37.Troudi A., Fethi M., El Asli M.S., Bolla J.M., Klibi N., Brunel J.M. Efficiency of a tetracycline-adjuvant combination against multidrug resistant Pseudomonas aeruginosa Tunisian clinical isolates. Antibiotics. 2020;9:919. doi: 10.3390/antibiotics9120919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang G., Brunel J.-M., Bolla J.-M., Van Bambeke F. The polyaminoisoprenyl potentiator NV716 revives old disused antibiotics against intracellular forms of infection by Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2021;65:e02028. doi: 10.1128/AAC.02028-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bisacchi G.S., Manchester J.I. A New-Class Antibacterial—Almost. Lessons in Drug Discovery and Development: A Critical Analysis of More than 50 Years of Effort toward ATPase Inhibitors of DNA Gyrase and Topoisomerase IV. ACS Infect. Dis. 2015;1:4–41. doi: 10.1021/id500013t. [DOI] [PubMed] [Google Scholar]
- 40.te Winkel J.D., Gray D.A., Seistrup K.H., Hamoen L.W., Strahl H. Analysis of Antimicrobial-Triggered Membrane Depolarization Using Voltage Sensitive Dyes. Front. Cell Dev. Biol. 2016;4:29. doi: 10.3389/fcell.2016.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borselli D., Blanchet M., Bolla J.-M., Muth A., Skruber K., Phanstiel O.I.V., Brunel J.M. Motuporamine Derivatives as Antimicrobial Agents and Antibiotic Enhancers against Resistant Gram-Negative Bacteria. Chembiochem. 2017;18:276–283. doi: 10.1002/cbic.201600532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naseem R., Wann K.T., Holland I.B., Campbell A.K. ATP regulates calcium efflux and growth in E. coli. J. Mol. Biol. 2009;391:42–56. doi: 10.1016/j.jmb.2009.05.064. [DOI] [PubMed] [Google Scholar]
- 43.Committee S.A. Comité de l’Antibiogramme de la Société Française de Microbiologie report 2003. Int. J. Antimicrob. Agents. 2003;21:364–391. doi: 10.1016/s0924-8579(03)00021-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.