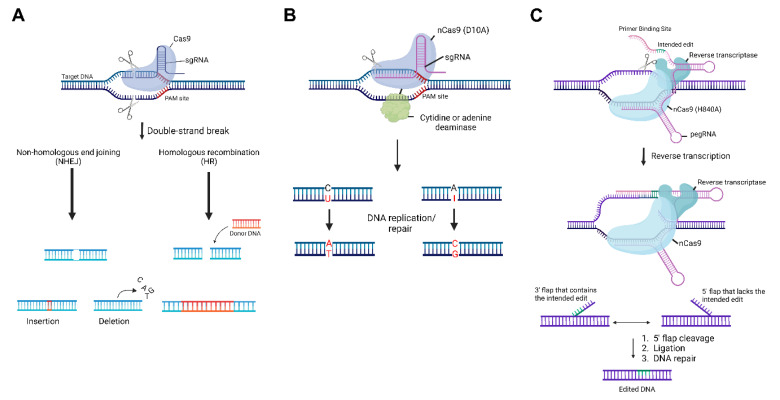
Figure 1.
Overview of the clustered regularly interspaced short palindromic repeats (CRISPR) toolbox. (A) Illustration of the original CRISPR/CRISPR-associated protein 9 (Cas9) System. Upon recognition of the target sequence, Cas9 will cleave both strands of deoxyribonucleic acid (DNA). This will result in two natural DNA repair pathways—non-homologous end joining and homologous recombination, which will lead to a small insertion or deletion, or a large insertion containing a donor DNA template, respectively. (B) Illustration of the base editing system. A Cas9 nickase (nCas9) is fused to a cytidine or adenine deaminase and upon nicking the single strand of DNA, the enzyme will deaminate the cytosine or adenine, leading to a uracil or inosine, respectively. Then, DNA replication or repair will recognize the change and lead to a permanent base pair conversion. (C) Illustration of the prime editing system. Prime editing utilizes a Cas9 nickase fused to a reverse transcriptase and a prime editing guide ribonucleic acid (pegRNA) that contains the spacer sequence, primer binding site, and the template containing the intended edit. After recognition and the single-strand nick, the primer binding site will allow for the exposed 3′-hydroxyl end of the nicked DNA strand to initiate the reverse transcription of the template. This results in an intermediate that includes two DNA flaps: a 3′ flap that contains the desired edit, and a 5′ flap that contains the unedited strand. After equilibration between the two flaps, cleavage, ligation, and DNA repair, the stably edited DNA sequence remains. PAM, protospacer adjacent motif; sgRNA, single guide ribonucleic acid.