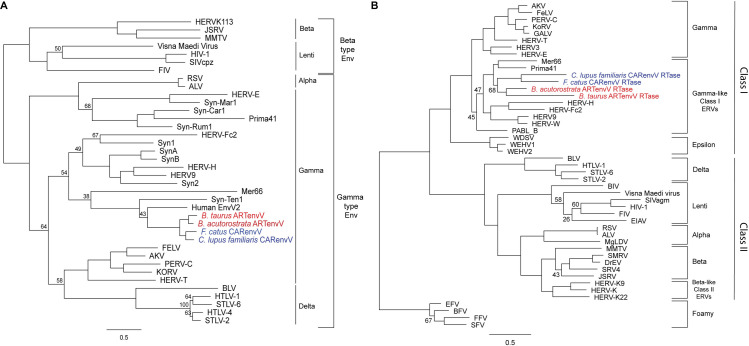
Fig 5. Phylogeny of CARenvV and ARTenvV proteins.
A. The predicted amino acid sequence of CARenvV (in blue) and ARTenvV (in red) from the indicated species together with the amino acid sequence of selected endogenous and exogenous retroviral Env proteins (S2 Table) were aligned, and a maximum likelihood tree was generated using RaxML with 500 bootstraps. The tree was midrooted. B. Amino acid sequence of the predicted RTase domains of CARenvV ERV (in blue) and ARTenvV ERV (in red) from the indicated species together with the amino acid sequence of the RTase domain from the indicated endogenous and exogenous retroviruses (S2 Table) were aligned and a maximum likelihood tree was generated using RaxML with 500 bootstraps. The tree was rooted using foamy viruses. Bootstrap values that are less than 70 are shown at the relevant nodes. Scale bar represents 0.5 amino acid substitutions per site.