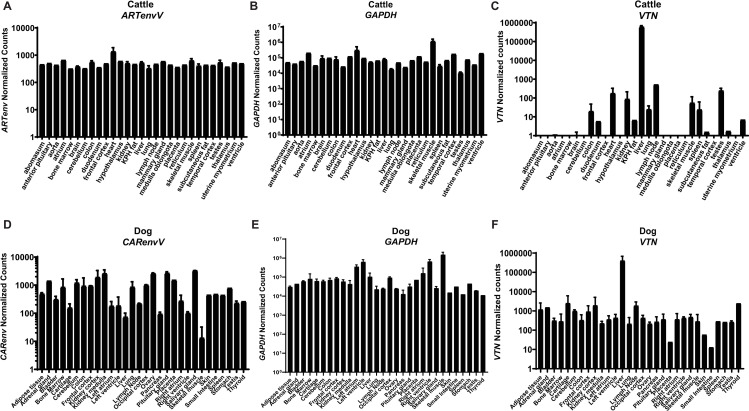
Fig 9. CARenvV and ARTenvV are expressed in a wide variety of tissues.
In silico analyses of RNA sequencing data from 29 tissues from cattle (B. taurus) and 30 tissues from domestic dog (C. lupus familiaris) are shown for A. cattle ARTenvV, B. cattle GAPDH, C. cattle VTN, D. dog CARenvV, E. dog GAPDH and F. dog VTN. The mapping accuracy of the RNA sequencing reads, and the normalization of the read counts was confirmed using as controls a liver specific gene (VTN) (C. and F.) or a housekeeping gene (GAPDH) (B. and E.). Raw RNA sequencing data extracted from SRA projects PRJNA314981, PRJNA78827, PRJNA379574, PRJNA177791 and PRJNA314981 were mapped to the cattle or dog genome assembly and the counts that map to the coding region of each gene were extracted and normalized using DEseq2. KPH: Kidney, pelvic and heart.